Abstract
The authors describe an electrochemical strategy for ultrasensitive and specific detection of microRNA (miRNA). It is based on both multicomponent nucleic acid enzyme (MNAzyme) amplification and rolling circle amplification (RCA). In the presence of target miRNAs, partial enzyme A (partzyme A) and partial enzyme B (partzyme B) are assembled to form active MNAzymes. Once formed, the MNAzymes catalyze the cleavage of the hairpin substrates to liberate biotinylated fragments which hybridized with the capture probes immobilized on a gold electrode. The RCA is then initiated to form a product that binds many detection probes. Finally, the amperometric signal (best acquired at a working voltage of 0.22 V vs. Ag/AgCl) is obtained by employing the streptavidinylated alkaline phosphatase as the enzyme. This biosensor has a 1.66 fM detection limit, and a dynamic range that extends from 10 fM to 1 nM. It displays specificity down to single mismatch discrimination of target miRNAs and good reproducibility. It was successfully applied to the determination of miRNA in total RNA samples extracted from human breast adenocarcinoma MCF-7 cells.

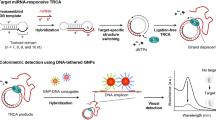
Cascade signal amplification strategy for microRNA electrochemical detection based on multicomponent nucleic acid enzyme (MNAzyme)-mediated rolling circle amplification. MCH: 6-mercapto-1-hexanol; BSA: bovine serum albumin; dNTP: deoxy-ribonucleoside triphosphate; ST-AP: streptavidin-alkaline phosphatase; α-NP: α-naphthyl phosphate





Similar content being viewed by others
References
He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531
Lujambio A, Lowe SW (2012) The microcosmos of cancer. Nature 482:347–355
Siegismund CS, Rohde M, Kühl U, Lassner D (2014) Multiparametric diagnostics of cardiomyopathies by microRNA signatures. Microchim Acta 181:1647–1653
Haramati S, Chapnik E, Sztainberg Y, Eilam R, Zwang R, Gershoni N, McGlinn E, Heiser PW, Wills AM, Wirguin I, Rubin LL, Misawa H, Tabin CJ, Jr RB, Chen A, Hornstein E (2010) MiRNA malfunction causes spinal motor neuron disease. Proc Natl Acad Sci 107:13111–13116
Esquela-Kerscher A, Slack FJ (2006) Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer 6:259–269
Hammond SM (2006) MicroRNA detection comes of age. Nat Methods 3:12–13
Liu L, Jiang S, Wang L, Zhang Z, Xie G (2015) Direct detection of microRNA-126 at a femtomolar level using a glassy carbon electrode modified with chitosan, graphene sheets, and a poly (amidoamine) dendrimer composite with gold and silver nanoclusters. Microchim Acta 182:77–84
Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES (2008) Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem 54:1696–1704
Varallyay E, Burgyan J, Havelda Z (2008) MicroRNA detection by northern blotting using locked nucleic acid probes. Nat Protoc 3:190–196
Nagl S, Schaeferling M, Wolfbeis OS (2005) Fluorescence analysis in microarray technology. Microchim Acta 151:1–21
Liu T, Chen X, Hong CY, XP X, Yang HH (2014) Label-free and ultrasensitive electrochemiluminescence detection of microRNA based on long-range self-assembled DNA nanostructures. Microchim Acta 181:731–736
Zhang D, Yan Y, Cheng W, Zhang W, Li Y, Ju H, Ding S (2013) Streptavidin-enhanced surface plasmon resonance biosensor for highly sensitive and specific detection of microRNA. Microchim Acta 180:397–403
Wang M, Yang Z, Guo Y, Wang X, Yin H, Ai S (2015) Visible-light induced photoelectrochemical biosensor for the detection of microRNA based on Bi2S3 nanorods and streptavidin on an ITO electrode. Microchim Acta 182:241–248
Zhang Y, Tang L, Yang F, Sun Z, Zhang G (2015) Highly sensitive DNA-based fluorometric mercury(II) bioassay based on graphene oxide and exonuclease III-assisted signal amplification. Microchim Acta 182:1535–1541
Wang M, Fu Z, Li B, Zhou Y, Yin H, Ai S (2014) One-step, ultrasensitive, and electrochemical assay of microRNAs based on T7 exonuclease assisted cyclic enzymatic amplification. Anal Chem 86:5606–5610
Zhang Y, Yan Y, Chen W, Cheng W, Li S, Ding X, Li D, Wang H, Ju H, Ding S (2015) A simple electrochemical biosensor for highly sensitive and specific detection of microRNA based on mismatched catalytic hairpin assembly. Biosens Bioelectron 68:343–349
Zhao X, Gong L, Zhang X, Yang B, Fu T, Hu R, Tan W, Yu R (2013) Versatile DNAzyme-based amplified biosensing platforms for nucleic acid, protein, and enzyme activity detection. Anal Chem 85:3614–3620
Freage L, Wang F, Orbach R, Willner I (2014) Multiplexed analysis of genes and of metal ions using enzyme/DNAzyme amplification machineries. Anal Chem 86:11326–11333
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510
Xiao Y, Rowe AA, Plaxco KW (2007) Electrochemical detection of parts-per-billion lead via an electrode-bound DNAzyme assembly. J Am Chem Soc 129:262–263
Mokany E, Bone SM, Young PE, Doan TB, Todd AV (2010) MNAzymes, a versatile new class of nucleic acid enzymes that can function as biosensors and molecular switches. J Am Chem Soc 132:1051–1059
Mokany E, Tan YL, Bone SM, Fuery CJ, Todd AV (2013) MNAzyme qPCR with superior multiplexing capacity. Clin Chem 59:419–426
Kolpashchikov DM (2007) A binary deoxyribozyme for nucleic acid analysis. Chembiochem 8:2039–2042
Jie G, Qin Y, Meng Q, Wang J (2015) Autocatalytic amplified detection of DNA based on a CdSe quantum dot/folic acid electrochemiluminescence energy transfer system. Analyst 140:79–82
Zagorovsky K, Chan WC (2013) A plasmonic DNAzyme strategy for point-of-care genetic detection of infectious pathogens. Angew Chem Int Ed 52:3168–3171
Ren K, Wu J, Ju H, Yan F (2015) Target-driven triple-binder assembly of MNAzyme for amplified electrochemical immunosensing of protein biomarker. Anal Chem 87:1694–1700
Gerasimova YV, Kolpashchikov DM (2010) Nucleic acid detection using MNAzymes. Chem Biol 2:104–106
Xu Z, Yin H, Tian Z, Zhou Y (2014) Electrochemical immunoassays for the detection the activity of DNA methyltransferase by using the rolling circle amplification technique. Microchim Acta 181:471–477
Chapin SC, Doyle PS (2011) Ultrasensitive multiplexed microRNA quantification on encoded gel microparticles using rolling circle amplification. Anal Chem 83:7179–7185
Cheng W, Ding L, Chen Y, Yan F, Ju H, Yin Y (2010) A facile scanometric strategy for ultrasensitive detection of protein using aptamer-initiated rolling circle amplification. Chem Commun 46:6720–6722
Cheng Y, Zhang X, Li Z, Jiao X, Wang Y, Zhang Y (2009) Highly sensitive determination of microRNA using target-primed and branched rolling-circle amplification. Angew Chem Int Ed 48:3268–3272
Hofmann S, Huang Y, Paulicka P, Kappel A, Katu HA, Keller A, Meder B, Stähler CF, Gumbrecht W (2015) Double-stranded ligation assay for the rapid multiplex quantification of microRNAs. Anal Chem 87:12104–12111
Hou T, Li W, Liu X, Li F (2015) Label-free and enzyme-free homogeneous electrochemical biosensing strategy based on hybridization chain reaction: a facile, sensitive, and highly specific MicroRNA assay. Anal Chem 87:11368–11374
Acknowledgments
This work was funded by the National Natural Science Foundation of China (81101638) and the Science and Technology Plan Project of Yu Zhong District of Chongqing (20150114).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author(s) declare that they have no competing interests.
Additional information
Jianru Yang and Min Tang contributed equally to this work.
Electronic supplementary material
ESM 1
(DOC 339 kb)
Rights and permissions
About this article
Cite this article
Yang, J., Tang, M., Diao, W. et al. Electrochemical strategy for ultrasensitive detection of microRNA based on MNAzyme-mediated rolling circle amplification on a gold electrode. Microchim Acta 183, 3061–3067 (2016). https://doi.org/10.1007/s00604-016-1958-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-016-1958-5