Abstract
Purpose
Accessible patient information sources are vital in educating patients about the benefits and risks of spinal surgery, which is crucial for obtaining informed consent. We aim to assess the effectiveness of a natural language processing (NLP) pipeline in recognizing surgical procedures from clinic letters and linking this with educational resources.
Methods
Retrospective examination of letters from patients seeking surgery for degenerative spinal disease at a single neurosurgical center. We utilized MedCAT, a named entity recognition and linking NLP, integrated into the electronic health record (EHR), which extracts concepts and links them to systematized nomenclature of medicine-clinical terms (SNOMED-CT). Investigators reviewed clinic letters, identifying words or phrases that described or identified operations and recording the SNOMED-CT terms as ground truth. This was compared to SNOMED-CT terms identified by the model, untrained on our dataset. A pipeline linking clinic letters to patient-specific educational resources was established, and precision, recall, and F1 scores were calculated.
Results
Across 199 letters the model identified 582 surgical procedures, and the overall pipeline after adding rules a total of 784 procedures (precision = 0.94, recall = 0.86, F1 = 0.91). Across 187 letters with identified SNOMED-CT terms the integrated pipeline linking education resources directly to the EHR was successful in 157 (78%) patients (precision = 0.99, recall = 0.87, F1 = 0.92).
Conclusions
NLP accurately identifies surgical procedures in pre-operative clinic letters within an untrained subspecialty. Performance varies among letter authors and depends on the language used by clinicians. The identified procedures can be linked to patient education resources, potentially improving patients’ understanding of surgical procedures.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Informed consent is the foundation of shared decision making in surgery. It is defined by the patient being aware of any material risks involved in a proposed treatment [1, 2]. Despite this, patient recall after spinal surgery consent is only 45% immediately after discussion and lack of informed consent is a commonly cited reason for medicolegal claims [3,4,5]. This is particularly challenging in spinal surgery, which carries a wide range of risks, and the perceived benefits may not match patient expectations [6]. It is well-recognized that providing additional information through videos and online resources when consenting patients for neurosurgery improves understanding of the procedure and its risks [7, 8]. However, the heterogeneity of spinal surgery means that it is difficult for clinicians to provide additional information that is both informative and relevant to the patient’s specific situation.
Electronic health records (EHR) have quickly become established in modern healthcare settings as a safe, practical, and time efficient alternative to paper records [9, 10]. The use of EHR generates a wealth of easily accessible data, albeit in an unstructured form which makes it highly time and resource intensive to analyze. Natural language processing (NLP) is a subfield of artificial intelligence, which interprets and contextualizes written language [11]. The clinic letter remains the cornerstone of physician-to-physician communication, and as such contains a high density of patient specific information. However, the free text nature of clinic letters means that the information may be underexploited by standard digital automation, which analyses formal labels and ticked boxes (structured data), rather than unstructured data such as natural language. It remains unclear if NLP can be used in the retrieval of unstructured data from clinic spine surgery letters to build a diagnostic and surgical treatment profile for an individual, which can then be utilized to provide automated patient specific and personalized educational resources. An effective and scalable NLP could unburden clinicians from compiling, synthesizing, and recommending educational resources themselves [11]. These duties may shift clinicians’ focus away from the patient and contribute to a high clerical workload which can lead to burnout [12]. The NLP is designed for clinicians as a clinical tool to enhance the consent process. This could result in a more personalized consent process, empowering patients with individualized educational resources for better comprehension of their diagnosis and proposed treatment. This, in turn, may facilitate more focused and higher-level discussions during the formal consent consultation.
The study objectives are to evaluate the utility of an NLP model in identifying procedures from outpatient clinic letters and using a simple pipeline link these procedures to patient information using degenerative spinal disease as an exemplar.
Methods
Study design and methods summary
This study was a retrospective analysis of clinical records of patients before an elective spinal operation at a single neurosurgical center in the United Kingdom.
This study utilized the Medical Concept Annotation Toolkit (MedCAT)—an NLP tool within the data retrieval software CogStack—to recognize descriptions of surgical procedures for degenerative spine disease and to identify the Systematized Nomenclature of Medicine—Clinical Terms (SNOMED-CT) terms. To test the performance of MedCAT, one preoperative clinic letter per patient was manually labelled for surgical procedures by identifying and recording procedures described in the written text. MedCAT was used to link extracted procedure concepts to SNOMED-CT terms, which was compared against the labelled ‘ground truth’.
A pipeline was established to link clinic letters to relevant educational resources through a patient-specific dashboard integrated into the EHR.
For each task above (SNOMED identification and patient resource retrieval), macro-averaged precision, recall, and F1 scores were calculated for this linkage, using the actual procedure received by the patient as the ground truth.
The methods are described in more detail below.
Participants
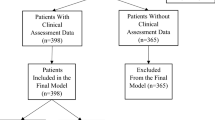
Patients were identified using a EHR system (Epic Caboodle, Epic Systems Corporation, Wisconsin, USA). The study population includes adult patients aged > 18 years old undergoing inpatient elective surgery for degenerative spine disease (both instrumented and non-instrumented), between January 2022 and June 2022. This study period was chosen to achieve a cohort target size of 200 patients who sequentially presented to the neurosurgical center. This number was chosen after discussion with a data engineer to provide sufficient data for evaluation of the NLP to be used, which had previously undergone extensive supervised and self-supervised validation on large biomedical datasets. Patients were excluded from the study if they had non-elective surgery or a non-degenerative cause of spinal disease, and if they did not have clinic letters saved to the EHR.
Data sources and measurements
Unstructured written information from clinic letters stored within the EHR were anonymized and extracted by the information retrieval platform CogStack [13]. The CogStack platform was developed specifically to comply with the strict data governance policies of the national health service (NHS) [14]. As there may have been several clinic letters prior to the surgery, it was decided that the most detailed clinic letter was extracted for each patient prior to surgery. In addition, demographic information–age at clinic, sex and diagnosis was recorded from the EHR.
MedCAT development
MedCAT is a ‘generalist’ medical NLP algorithm that uses machine learning (ML) to perform named entity recognition (NER). The goal of the NER is to extract information about specific types of entities (diagnoses, symptoms, or procedures) in written text that it can link to SNOMED-CT terms.
MedCAT was trained through a combination of supervised and self-supervised ML techniques, and the evaluation of concept recognition was conducted using various publicly accessible datasets, including MedMentions [15], ShARe/CLEF 2014 Task 2 [16], and MIMIC-III [17]. Additional validation was performed using electronic health records (EHR) from three major University hospitals in the UK [18]. Moreover, MedCAT underwent supervised learning in previous neurosurgical projects focusing on hydrocephalus [19] and skull base neurosurgery.
Pipeline evaluation
The MedCAT analysis was carried out in distinct stages (Fig. 1). Firstly, two neurosurgeons (JP, JB) reviewed the last three clinic letters prior to surgery for patients (N = 199) undergoing elective degenerative spine surgery and identified the most detailed of these clinic letters to label the documented spinal surgical procedure. In each clinic letter, the neurosurgeons labelled any descriptions of a surgical procedure in the letter (multi-label) and the single proposed surgical procedure that was offered to the patient. Secondly, MedCAT was tested on its ability correctly link descriptions of surgical procedures to the correct SNOMED-CT term from a list agreed upon by the neurosurgical doctors (Supplement 1). Multiple SNOMED-CT terms may be identified from each clinic letter due to clinicians repeating procedure terms or describing other procedures (e.g., discussing alternative options). This was compared against the surgical procedures identified by the neurosurgical doctors. True positive, false positive and false negative values were recorded for each SNOMED-CT concept. The NLP pipeline was optimized by adding in ‘rules’ (Supplement 2) to ensure that synonyms of procedures were identified under the relevant SNOMED CT term. Thirdly, a list of trusted educational resources was manually compiled from british association of spinal surgeons (BASS) and NHS trusts websites. The surgical procedures extracted from the NLP were then used to retrieve patient-information resources, that were linked electronically to the clinic letter on each patient’s EHR.
Data analysis
The performance of the MedCAT model was evaluated during three separate stages: base MedCAT, MedCAT refined with added rules, and resource linkage. At each stage we calculated the precision, recall and F1 score of the model. Precision measures the proportion of true positive predictions out of all the positive predictions made by the model. In the context of MedCAT, it represents the accuracy of the model in identifying a spinal procedure. Recall measures the proportion of true positive predictions out of all the actual positive instances in the dataset. In the context of MedCAT, it represents how well the model captures all the spinal procedures mentioned in the clinic letters. The F1 score is the harmonic mean of precision and recall. It provides a balance between precision and recall, giving a single value that summarizes the model’s overall performance [20].
Results
Summary of data
Between January and June 2022, 199 patients who underwent surgery for degenerative spine disease had their clinic letters analyzed. The most common diagnoses were lumbar degenerative disc disease (34.2%) and cervical degenerative disc disease (20.1%). The basic demographic information of this cohort shown in Table 1.
Concept extraction
Initially, the MedCAT tool alone was applied to the clinical letters without additional processes in the NLP pipeline. In 187 (94%) letters the base MedCAT model identified 582 SNOMED-CT surgical procedure terms with a macro-average precision = 0.93, recall = 0.86, and F1 = 0.88 (supplement 2). In 12 letters, written text in the clinic letter did not use any text that could be linked to the prespecified list of SNOMED-CT terms or synonyms of procedures.
Using the base MedCAT model, there were many terms that were not identified as SNOMED-CT terms due to variations in written language. In some cases, the official SNOMED-CT term varied hugely from the term used to describe the procedure in clinic letters. For example, ‘ACDF’ was commonly used in clinic letters to describe the SNOMED-CT term ‘cervical arthrodesis by anterior technique’. Therefore, rules were applied to the MedCAT model to refine the pipeline and improve the uptake of SNOMED-CT terms (Supplement 2).
Pipeline refinement
The addition of ‘rules’ to identify synonyms added a further 202 surgical procedures. The optimized MedCAT model identified 784 surgical procedures, the overall macro average performance metrics were precision = 0.98, recall = 0.86, and F1 = 0.91 (Supplement 3).
Linking extracted terms with educational resources
A proof-of-concept alerting dashboard, integrated into the EHR, was used offline to surface patients who have been identified by MedCAT as having a spinal procedure (Fig. 2). Clinicians were able to accept or reject the alert on the dashboard, and this would in turn trigger a sequence of automated tasks that inserts/attaches relevant resources into patient’s next letter/correspondence. A proposed pipeline for sending educational resources to patients is shown in Fig. 3.
The model successfully linked extracted concepts from the letter to a specific educational resource in 157 (78%) of the 202 mutually exclusive spinal procedures performed. Overall, the macro-averages were precision = 0.99, recall = 0.87, F1 = 0.92. The precision, recall and F1 scores for this linking was calculated for each procedure (Table 2).
Discussion
Principal findings
This study evaluated the use of an NLP model to extract SNOMED-CT terms from clinic letters. By adding simple additional processes to the pipeline, it was possible to link the identified surgical procedure with specific educational resources for patients.
Firstly, the base MedCAT model performed well without dedicated training on the dataset as indicated by precision = 0.93, recall = 0.86, and F1 = 0.88. Following refinement of the pipeline to identify synonyms of the SNOMED-CT terms, the precision of MedCAT model improved, but the recall score remained unchanged (precision = 0.98, recall = 0.86, and F1 = 0.91). This is to say that the accuracy of the MedCAT model in identifying SNOMED-CT terms improved, but the overall number of SNOMED-CT terms it identified remained unchanged. These findings collectively affirm MedCAT’s potential as a valuable tool for extracting and categorizing medical information from clinic letters, offering promising prospects for integration into clinical practice. The model's capacity to incorporate rules dynamically through a user interface, improving precision in real-time, renders it especially attractive for supporting data collection processes in healthcare settings.
Secondly, dedicated training is not required for NLP models that have been previously trained on a large volume of clinical documents, even when applied to a highly-specialist area such as spinal neurosurgery. The MedCAT model was trained with supervised and self-supervised ML on several publicly available patient databases with additional validation using EHR from three major University hospitals in the UK [18]. However, the MedCAT model did not undergo any prior training specific to spinal neurosurgery, which indicates that ‘generalist’ NLP may perform well in biomedical settings without prior training. This supports the use of a widely available Trusted Research Environments (TSE), such as CogStack, that have integrated NLP tools and can be distributed among multiple different healthcare settings, as opposed to developing multiple TSEs for each separate setting.
Thirdly, while the MedCAT model performed well at linking written text to SNOMED-CT concepts it performed less well at retrieving relevant educational resources for patients. The MedCAT model couldn’t distinguish between past spinal surgeries mentioned in clinic letters and upcoming proposed spinal procedures. Other, errors occurred when letters contained the process of discussion when considering multiple surgical options, the MedCAT model would identify each of the procedures mentioned as a separate proposed spinal procedure, despite only a single operative plan being decided upon. The result was that 78% of letters could be linked directly to a relevant educational resource. We acknowledge that clinicians currently have a higher performance to provide educational resources. However, the current practice involves clinicians manually searching the internet for resources and printing them during consultations, which poses several challenges: (1) dependence on clinicians’ memory, (2) time-consuming, (3) lacks scalability, and (4) internet resources may be variable in quality and subject to change. Further development is required of the model in interpreting letters with multiple procedural names and an overseeing clinician is recommended to ‘accept’ or ‘reject’ recommended resources, to ensure their relevance. Once functional, this clinical tool will be highly scalable to other healthcare settings, relieving clinicians of the burden of remembering and manually sourcing trusted educational resources.
Fourthly, we demonstrated that use of NLP models based on NER linking with SNOMED-CT nomenclature is limited as it does not reflect the way diagnoses are communicated to patients in a clinical setting. We found that a higher false negative rate occurred for decompression of the lumbar spine because the MedCAT NLP identified the verb ‘decompress’ as a procedural name–decompression of lumbar spine. As decompress is common language to use when describing a large variety of spinal operations, it led to a higher false negative rate. In addition, neurosurgeons used multiple different ways to describe the same spinal procedure. To account for this multiple ‘rules’ were created for each surgical procedure to link synonyms and descriptions to SNOMED-CT terms and prevent false negatives. Despite this, 12 letters used descriptive of procedures that could not be linked to SNOMED-CT terms. The heterogeneous descriptions of surgical procedures in spinal surgery represent a well-recognized problem, lacking a systematized language for effective communication. The resulting inconsistent and varied nomenclature can lead to confusion amongst patients, clinicians, and researchers [21]. A recent paper systematically analyzed the nomenclature used in the literature to describe a lateral interbody fusion procedure and identified 72 distinct ways [22]. There have been recent attempts to standardize the use of terms [23], but our study has indicated that it is an ongoing problem. We acknowledge that large language models exhibit potential advantages in interpreting text, particularly in domains with extensive language variations, such as spinal clinic letters. However, it is crucial to recognize that, at present, the integration of these large language models poses challenges in complying with the stringent data governance policies of the NHS and other healthcare systems.
Strengths and limitations of the study
In this study, we demonstrate the value of NLP in a preoperative setting to identify surgical procedures, which can be linked with educational resources. The use case described in this study is completely novel and has wide-reaching implications for patients. The MedCAT NLP used required no dedicated training, is low cost and will be widely available to any healthcare system that uses EHR.
The data was collected from a single center, and the results may not be generalizable to other centers which clinical letters are not written in English or EHR are not available. Another limitation is that the NLP was used ‘offline’ and it is unclear if the tool will have clinical benefit for patients. The MedCAT tool now needs to be implemented into clinical practice to investigate the extent it will benefit patient’s understanding of surgical procedures.
Conclusions
This study demonstrates the ability for an NLP algorithm, with no prior task specific training to identify surgical procedures from pre-operative clinical letters with high precision, which can be linked with specific patient education resources. Errors in the model arose due to variations in terminology used to describe spinal procedures and the model being unable to differentiate previous from future surgical procedures. Further development the NLP algorithm may lead to improved performance when linking surgical procedures with relevant educational resources. This study clinical implications for improving patient understanding of surgical procedures and empowering them engage in shared decision making.
References
Sokol DK (2015) Update on the UK law on consent. BMJ 350:1481. https://doi.org/10.1136/bmj.h1481
House of Lords - Chester (Respondent) v. Afshar (Appellant). https://publications.parliament.uk/pa/ld200304/ldjudgmt/jd041014/cheste-1.htm. Accessed 28 Aug 2022
Saigal R, Clark AJ, Scheer JK (2015) Adult spinal deformity patients recall fewer than 50% of the risks discussed in the informed consent process preoperatively and the recall rate worsens significantly in the postoperative period. Spine (Phila Pa 1976) 40:1079–1085. https://doi.org/10.1097/BRS.0000000000000964
Machin JT, Hardman J, Harrison W et al (2018) Can spinal surgery in England be saved from litigation: a review of 978 clinical negligence claims against the NHS. Eur Spine J 27:2693–2699. https://doi.org/10.1007/s00586-018-5739-1
Renovanz M, Haaf J, Nesbigall R et al (2019) Information needs of patients in spine surgery: development of a question prompt list to guide informed consent consultations. Spine J 19:523–531. https://doi.org/10.1016/j.spinee.2018.08.015
Fahey N, Patel V, Rosseau G (2014) A comparative analysis of online education resources for patients undergoing endoscopic transsphenoidal surgery. World Neurosurg 82:E671–E675. https://doi.org/10.1016/j.wneu.2014.09.014
Marcus HJ, Jain A, Grieve J, Dorward NL (2018) Informed consent for patients undergoing transsphenoidal excision of pituitary adenoma: development and evaluation of a procedure-specific online educational resource. World Neurosurg 118:e933–e937. https://doi.org/10.1016/j.wneu.2018.07.102
Lee YS, Cho DC, Sung JK et al (2020) The effect of an educational and interactive informed consent process on patients with cervical spondylotic myelopathy caused by ossification of the posterior longitudinal ligament. Spine (Phila Pa 1976) 45:193–200. https://doi.org/10.1097/BRS.0000000000003223
Lin SC, Jha AK, Adler-Milstein J (2018) Electronic health records associated with lower hospital mortality after systems have time to mature. Health Aff 37:1128–1135. https://doi.org/10.1377/hlthaff.2017.1658
Holmgren AJ, Pfeifer E, Manojlovich M, Adler-Milstein J (2016) A novel survey to examine the relationship between health it adoption and nurse-physician communication. Appl Clin Inform 07:1182–1201
Hashimoto DA, Rosman G, Rus D, Meireles OR (2018) Artificial intelligence in surgery: promises and perils. Ann Surg 268:70–76. https://doi.org/10.1097/SLA.0000000000002693
Patel RS, Bachu R, Adikey A et al (2018) Factors related to physician burnout and its consequences: a review. Behav Sci (Basel, Switzerland). https://doi.org/10.3390/bs8110098
Noor K, Roguski L, Handy A et al (2021) Deployment of a free-text analytics platform at a UK National Health Service Research Hospital: CogStack at university college london hospitals. JMIR Med Inform 10:e38122. https://doi.org/10.2196/38122
Jackson R, Kartoglu I, Stringer C et al (2018) CogStack-experiences of deploying integrated information retrieval and extraction services in a large national health service foundation trust hospital. BMC Med Inform Decis Mak 18:47. https://doi.org/10.1186/s12911-018-0623-9
Mohan S, Li D, Zuckerberg C (2019) MedMentions: a large biomedical corpus annotated with umls concepts
Mowery DL, Velupillai S, South BR, et al (2013) Task 2: ShARe/CLEF eHealth evaluation lab 2014
Johnson AEW, Pollard TJ, Shen L et al (2016) MIMIC-III, a freely accessible critical care database. Sci Data 3:160035. https://doi.org/10.1038/sdata.2016.35
Kraljevic Z, Searle T, Shek A et al (2021) Multi-domain clinical natural language processing with MedCAT: the medical concept annotation toolkit. Artif Intell Med. https://doi.org/10.1016/j.artmed.2021.102083
Funnell JP, Noor K, Khan DZ et al (2022) Characterization of patients with idiopathic normal pressure hydrocephalus using natural language processing within an electronic healthcare record system. J Neurosurg. https://doi.org/10.3171/2022.9.jns221095
Erickson BJ, Kitamura F (2021) Magician’s corner: 9. Performance metrics for machine learning models. Radiol Artif Intell 3(3):e200126. https://doi.org/10.1148/ryai.2021200126
Yucesoy K, Sonntag VK (2000) Terminology confusion in spinal surgery: laminotomy, laminoplasty, laminectomy. J Neurosurg 92:371. https://doi.org/10.3171/jns.2000.92.2.0371
Turlip R, Ahmad HS, Ghenbot YG et al (2023) Characterizing and improving nomenclature for reporting lumbar interbody fusion techniques. World Neurosurg 175:e134–e140. https://doi.org/10.1016/j.wneu.2023.03.040
McCloskey K, Singh S, Ahmad HS et al (2023) Standardizing lumbar interbody fusion nomenclature. Clin Spine Surg 36:217–219. https://doi.org/10.1097/BSD.0000000000001425
Acknowledgements
No specific funding was received for this piece of work. This study has been supported by the National Institute for Health Research (NIHR) University College London Hospitals Biomedical Research Centre, in particular by the NIHR UCLH/UCL BRC Clinical and Research Informatics Unit. JB, JP, JPF, DZK, NN, SS, SCW & HJM are supported by the Wellcome (203145Z/16/Z) EPSRC (NS/A000050/1) Centre for Interventional and Surgical Sciences, University College London. DZK is supported by a NIHR Academic Clinical Fellowship. KN, RJBD, & HJM are supported by the NIHR Biomedical Research Centre, University College London. CHK is supported by the Cleveland Clinic London MPhil/PhD fellowship. RJBD is additionally supported by the following: (1) NIHR Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, London, UK; (2) Health Data Research UK, which is funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation and Wellcome Trust; (3) The BigData@Heart Consortium, funded by the Innovative Medicines Initiative-2 Joint Undertaking under Grant agreement No. 116074. This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation program and EFPIA; it is chaired by DE Grobbee and SD Anker, partnering with 20 academic and industry partners and ESC; (4) NIHR Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London; (5) UK Research and Innovation London Medical Imaging & Artificial Intelligence Centre for Value Based Healthcare; (6) NIHR Applied Research Collaboration South London (NIHR ARC South London) at King’s College Hospital NHS Foundation Trust.
Author information
Authors and Affiliations
Contributions
JB, JP, RJBD, JPF, CHK, DZK, NN, KN, DR, SS, SCW, PS and HJM contributed to conceiving and designing the study. JB, JP and DR contributed to data extraction, curation, and analysis. RJBD and KN contributed to model design and development. JB, JP, JPF, DZK, NN, SS, SCW and HJM drafted the manuscript. RJBD, PS, and HJM provided supervision of the study. All authors were involved in the writing and approval of the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was registered as part of a service evaluation within University College London Hospitals and approved by the Clinical Governance Committee. Informed consent was not required for this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Booker, J., Penn, J., Noor, K. et al. Early evaluation of a natural language processing tool to improve access to educational resources for surgical patients. Eur Spine J (2024). https://doi.org/10.1007/s00586-024-08315-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00586-024-08315-5