Abstract
Background
The association between the evolution of hepatitis B virus (HBV) quasispecies and the development of hepatocellular carcinoma (HCC) is unknown.
Methods
We used deep sequencing to examine the dynamics of HBV quasispecies and their relationship to HCC development. Thirty-two chronic hepatitis B (CHB) patients with HCC (HCC group) and 32 matched CHB patients without HCC (controls) were recruited. Fourteen patients from each group had serial sera available up to 9 years before the time of the present study. Deep sequencing of the HBV pre-S regions was performed. HBV quasispecies complexity, diversity, and intrapatient prevalence of pre-S deletions/mutations were analyzed.
Results
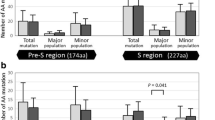
Compared with control patients, HCC patients had a significant greater quasispecies complexity (p = 0.04 at the nucleotide level), greater diversity (p = 0.004 and 0.009 at the nucleotide level and the amino acid level respectively), and a trend of greater complexity at the amino acid level (p = 0.065). HCC patients had a higher intrapatient prevalence of pre-S deletions and point mutations (at codons 4, 27, and 167) compared with the control patients (all p < 0.05). Longitudinal observation in the sera of 14 HCC patients showed that quasispecies complexity (p = 0.027 and 0.024 at the nucleotide level and the amino acid level respectively) and diversity (p = 0.035 and 0.031 at the nucleotide level and the amino acid level respectively) increased as the disease progressed to HCC.
Conclusions
Increased HBV quasispecies complexity and diversity in the pre-S region, probably reflecting enhanced virus–host interplay, was associated with disease progression from CHB to HCC.




Similar content being viewed by others
References
Fattovich G, Bortolotti F, Donato F. Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J Hepatol. 2008;48:335–52.
Domingo E, Gomez J. Quasispecies and its impact on viral hepatitis. Virus Res. 2007;127:131–50.
Fishman SL, Branch AD. The quasispecies nature and biological implications of the hepatitis C virus. Infect Genet Evol. 2009;9:1158–67.
Chevaliez S, Rodriguez C, Pawlotsky JM. New virologic tools for management of chronic hepatitis B and C. Gastroenterology. 2012;142(1303–1313):e1.
Ciftci S, Keskin F, Cakiris A, et al. Analysis of potential antiviral resistance mutation profiles within the HBV reverse transcriptase in untreated chronic hepatitis B patients using an ultra-deep pyrosequencing method. Diagn Microbiol Infect Dis. 2014;79:25–30.
Ko SY, Oh HB, Park CW, et al. Analysis of hepatitis B virus drug-resistant mutant haplotypes by ultra-deep pyrosequencing. Clin Microbiol Infect. 2012;18:E404–11.
Rodriguez C, Chevaliez S, Bensadoun P, et al. Characterization of the dynamics of hepatitis B virus resistance to adefovir by ultra-deep pyrosequencing. Hepatology. 2013;58:890–901.
Lin CL, Liu CH, Chen W, et al. Association of pre-S deletion mutant of hepatitis B virus with risk of hepatocellular carcinoma. J Gastroenterol Hepatol. 2007;22:1098–103.
Yeung P, Wong DKH, Lai CL, et al. Association of hepatitis B virus pre-S deletions with the development of hepatocellular carcinoma in chronic hepatitis B. J Infect Dis. 2011;203:646–54.
Zhang AY, Lai CL, Huang FY, et al. Evolutionary changes of hepatitis B virus pre-S mutations prior to development of hepatocellular carcinoma. PLoS One. 2015;10:e0139478.
Kozich JJ, Westcott SL, Baxter NT, et al. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–20.
Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41.
Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9.
Lim SG, Cheng Y, Guindon S, et al. Viral quasi-species evolution during hepatitis be antigen seroconversion. Gastroenterology. 2007;133:951–8.
Wu S, Imazeki F, Kurbanov F, et al. Evolution of hepatitis B genotype C viral quasi-species during hepatitis B e antigen seroconversion. J Hepatol. 2011;54:19–25.
Abe K, Thung SN, Wu HC, et al. Pre-S2 deletion mutants of hepatitis B virus could have an important role in hepatocarcinogenesis in Asian children. Cancer Sci. 2009;100:2249–54.
Hsieh YH, Su IJ, Wang HC, et al. Pre-S mutant surface antigens in chronic hepatitis B virus infection induce oxidative stress and DNA damage. Carcinogenesis. 2004;25:2023–32.
Yin J, Xie J, Zhang H, et al. Significant association of different preS mutations with hepatitis B-related cirrhosis or hepatocellular carcinoma. J Gastroenterol. 2010;45:1063–71.
Chen CH, Changchien CS, Lee CM, et al. Combined mutations in pre-s/surface and core promoter/precore regions of hepatitis B virus increase the risk of hepatocellular carcinoma: a case-control study. J Infect Dis. 2008;198:1634–42.
Gallina A, De Koning A, Rossi F, et al. Intracellular retention of hepatitis B virus surface protein mutants devoid of amino-terminal pre-S1 sequences. J Gen Virol. 1994;75(Pt 2):449–55.
Pollicino T, Amaddeo G, Restuccia A, et al. Impact of hepatitis B virus (HBV) preS/S genomic variability on HBV surface antigen and HBV DNA serum levels. Hepatology. 2012;56:434–43.
Cao L, Wu C, Shi H, et al. Coexistence of hepatitis B virus quasispecies enhances viral replication and the ability to induce host antibody and cellular immune responses. J Virol. 2014;88:8656–66.
Van den Hoecke S, Verhelst J, Vuylsteke M, et al. Analysis of the genetic diversity of influenza a viruses using next-generation DNA sequencing. BMC Genomics. 2015;16:79.
Quail MA, Smith M, Coupland P, et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, AY., Lai, CL., Huang, FY. et al. Deep sequencing analysis of quasispecies in the HBV pre-S region and its association with hepatocellular carcinoma. J Gastroenterol 52, 1064–1074 (2017). https://doi.org/10.1007/s00535-017-1334-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00535-017-1334-1