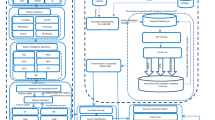
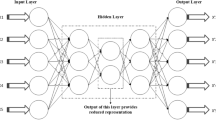
Abstract
Cancer is the major leading reason of death around the world. However, the early identification and prediction of a cancer type is very critical for patient’s health. Recently, microarray gene expression data was utilized for efficient and early diagnosis of cancer. Previous work shows that microarray data has two major issues which are high dimensionality and small sample size. Several researchers have analyzed and evaluated the cancer classification problem using different statistical and machine learning-based approaches but there are still some issues with these approaches that make cancer classification a nontrivial task. Such as, the inability of certain machine learning algorithms to use unstructured data has limited their utility in the cancer classification process. Convolutional neural networks are proven to very suitable to analyze variety of unstructured data. This ability allowed the deep learning algorithms to play a vibrant part in early detection of cancer through data classification. In this research, a hybrid deep learning model based on Laplacian Score-Convolutional Neural Network (LS-CNN) is employed for the classification of given cancer’s data. The performance of the proposed system was evaluated on 10 different benchmark datasets using various performance measurement metrics such as accuracy and confusion matrix. The experimental results conclude that proposed LS-CNN model outperformed compared to traditional machine learning and recently used deep learning approaches.





Similar content being viewed by others
References
NIH (2019) National Cancer Institute (NCI), cancer statistics. Available from: https://www.cancer.gov/. Accessed 23 April 2019
World Health Organization, Cancer (2018) Available from: https://www.who.int/news-room/fact-sheets/detail/cancer. Accessed 23 April 2019
Babu M, Sarkar K (2016) A comparative study of gene selection methods for cancer classification using microarray data. In: 2016 second international conference on research in computational intelligence and communication networks (ICRCICN). IEEE
Arslan MT, Kalinli A (2016) A comparative study of statistical and artificial intelligence based classification algorithms on central nervous system cancer microarray gene expression data. Int J Intell Syst Appl Eng. https://doi.org/10.18201/ijisae.267094
Bolón-Canedo V, Sánchez-Marono N, Alonso-Betanzos A, Benítez JM, Herrera F (2014) A review of microarray datasets and applied feature selection methods. Inf Sci 282:111–135
Hu H, Niu Z, Bai Y, Tan X (2015) Cancer classification based on gene expression using neural networks. Genet Mol Res 14:17605–17611
Bhola A, Tiwari AK (2015) Machine learning based approaches for cancer classification using gene expression data. Mach Learn Appl Int J 2(3/4):01–12
Singh RK, Sivabalakrishnan M (2015) Feature selection of gene expression data for cancer classification: a review. Proc Comput Sci 50:52–57
Gölcük G (2017) Cancer classification using gene expression data with deep learning. Paper presented at Department of Electronics, Informatics and Bioengineering Polytechnic University of Milan, Italy, 20 Dec 2017. http://hdl.handle.net/10589/138427
Khan MZ, Harous S, Hassan SU, Khan MUG, Iqbal R, Mumtaz S (2019) Deep unified model for face recognition based on convolution neural network and edge computing. IEEE Access 7:72622–72633
Guillen P, Ebalunode J (2016) Cancer classification based on microarray gene expression data using deep learning. In: 2016 international conference on computational science and computational intelligence (CSCI). IEEE
Bhat RR, Viswanath V, Li X (2017) DeepCancer: detecting cancer via deep generative learning through gene expressions. In: 2017 IEEE 15th international conference on dependable, autonomic and secure computing, 15th international conference on pervasive intelligence and computing, 3rd international conference on big data intelligence and computing and cyber science and technology congress (DASC/PiCom/DataCom/CyberSciTech). IEEE
Danaee P, Ghaeini R, Hendrix DA (2017) A deep learning approach for cancer detection and relevant gene identification. In: Pacific symposium on biocomputing 2017. World Scientific
Wenyan Z, Xuewen L, Jingjing W (2017) Feature selection for cancer classification using microarray gene expression data. Biostat Biom Open Access J 1(2):555557
Dang S, Wen M, Mumtaz S, Li J, Li C (2020) Enabling multi-carrier relay selection by sensing fusion and cascaded ANN for intelligent vehicular communications. IEEE Sens J. https://doi.org/10.1109/JSEN.2020.2986322
Matsubara T, Ochiai T, Hayashida M, Akutsu T, Nacher JC (2019) Convolutional neural network approach to lung cancer classification integrating protein interaction network and gene expression profiles. J Bioinform Comput Biol 17(03):1940007
Hamena S, Meshoul S (2018) Multi-class classification of gene expression data using deep learning for cancer prediction. Int J Mach Learn Comput 8(5):454–459
Luque-Baena R, Urda D, Subirats J, Franco L, Jerez J (2013) Analysis of cancer microarray data using constructive neural networks and genetic algorithms. In: Proceedings of the IWBBIO, international work-conference on bioinformatics and biomedical engineering
Natarajan A, Ravi T (2014) A survey on gene feature selection using microarray data for cancer classification. Int J Comput Sci Commun (IJCSC) 5(1):126–129
Kong Y, Yu T (2018) A deep neural network model using random forest to extract feature representation for gene expression data classification. Sci Rep 8(1):16477
Kumar M, Rath NK, Swain A, Rath SK (2015) Feature selection and classification of microarray data using MapReduce based ANOVA and K-nearest neighbor. Proc Comput Sci 54:301–310
Iqbal MS, Ahmad I, Bin L, Khan S, Rodrigues JJ (2020) Deep learning recognition of diseased and normal cell representation. Trans Emerg Telecommun Technol. https://doi.org/10.1002/ett.4017
Lyu B, Haque A (2018) Deep learning based tumor type classification using gene expression data. In: Proceedings of the 2018 ACM international conference on bioinformatics, computational biology, and health informatics. ACM
Khalifa NEM, Taha MHN, Ali DE, Slowik A, Hassanien AE (2020) Artificial intelligence technique for gene expression by tumor RNA-seq data: a novel optimized deep learning approach. IEEE Access 8:22874–22883
Khan S, Muhammad K, Mumtaz S, Baik SW, de Albuquerque VHC (2019) Energy-efficient deep CNN for smoke detection in foggy IoT environment. IEEE Internet Things J 6(6):9237–9245
Reena G (2011) A survey of human cancer classification using micro array data. Int J Comput Technol Appl 2(5):1523–1533. http://www.ijcta.com/vol2issue5-page3.php
Joseph M, Devaraj M, Leung CK (2019) DeepGx: deep learning using gene expression for cancer classification. In: 2019 IEEE/ACM international conference on advances in social networks analysis and mining (ASONAM). IEEE
Mostavi M, Chiu Y-C, Huang Y, Chen Y (2020) Convolutional neural network models for cancer type prediction based on gene expression. BMC Med Genom 13:1–13
Vimaladevi M, Kalaavathi B (2014) A microarray gene expression data classification using hybrid back propagation neural network. Genetika 46(3):1013–1026
Zeebaree DQ, Haron H, Abdulazeez AM (2018) Gene selection and classification of microarray data using convolutional neural network. In: 2018 international conference on advanced science and engineering (ICOASE). IEEE
Mao Z, Cai W, Shao X (2013) Selecting significant genes by randomization test for cancer classification using gene expression data. J Biomed Inform 46(4):594–601
Zhong W (2014) Feature selection for cancer classification using microarray gene expression data. University of Calgary, Calgary
Tabares-Soto R, Orozco-Arias S, Romero-Cano V, Bucheli VS, Rodríguez-Sotelo JL, Jiménez-Varón CF (2020) A comparative study of machine learning and deep learning algorithms to classify cancer types based on microarray gene expression data. PeerJ Comput Sci 6:e270
Salem H, Attiya G, El-Fishawy N (2017) Classification of human cancer diseases by gene expression profiles. Appl Soft Comput 50:124–134
Liu J, Wang X, Cheng Y, Zhang L (2017) Tumor gene expression data classification via sample expansion-based deep learning. Oncotarget 8(65):109646
Lee K, Man Z, Wang D, Cao Z (2013) Classification of bioinformatics dataset using finite impulse response extreme learning machine for cancer diagnosis. Neural Comput Appl 22(3–4):457–468
Wu Q, Boueiz A, Bozkurt A, Masoomi A, Wang A, DeMeo DL, Weiss ST, Qiu W (2018) Deep learning for predicting disease status using genomic data. PeerJ Preprints
Liu Y, Zhang N, He Y, Lun L (2015) Prediction of core cancer genes using a hybrid of feature selection and machine learning methods. Genet Mol Res 14(3):8871–8882
He X, Cai D, Niyogi P (2006) Laplacian score for feature selection. In: Advances in neural information processing systems
Mandal S, Banerjee I (2015) Cancer classification using neural network. Int J Emerg Eng Res Technol 3(7):172–178
Liu B, Wei Y, Zhang Y, Yang Q (2017) Deep neural networks for high dimension, low sample size data. In: IJCAI
Kim B-H, Yu K, Lee PC (2020) Cancer classification of single-cell gene expression data by neural network. Bioinformatics 36(5):1360–1366
Smolander J (2016) Deep learning classification methods for complex disorders
Fakoor R, Ladhak F, Nazi A, Huber M (2013) Using deep learning to enhance cancer diagnosis and classification. In: Proceedings of the international conference on machine learning. ACM, New York, USA
Zhou W, Dickerson JA (2014) A novel class dependent feature selection method for cancer biomarker discovery. Comput Biol Med 47:66–75
Liu J, Cai W, Shao X (2011) Cancer classification based on microarray gene expression data using a principal component accumulation method. Sci China Chem 54(5):802–811
Nagpal A, Singh V (2018) Identification of significant features using random forest for high dimensional microarray data. J Eng Sci Technol 13(8):2446–2463
Ram M, Najafi A, Shakeri MT (2017) Classification and biomarker genes selection for cancer gene expression data using random forest. Iran J Pathol 12(4):339
Acknowledgements
This work has been partially supported by FCT/MCTES through national funds and when applicable co-funded EU funds under the Project UIDB/50008/2020; and by Brazilian National Council for Scientific and Technological Development (CNPq) via Grant No. 309335/2017-5 and HEC Pakistan under NRPU funded Project No. 6338.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shah, S.H., Iqbal, M.J., Ahmad, I. et al. Optimized gene selection and classification of cancer from microarray gene expression data using deep learning. Neural Comput & Applic (2020). https://doi.org/10.1007/s00521-020-05367-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00521-020-05367-8