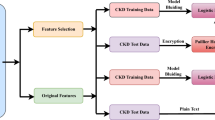
Abstract
Tuberculosis is one of the leading causes of millions of deaths across the world, mainly due to growth of drug-resistant strains. Anti-tubercular peptides may facilitate an alternate way to combat antibiotic tolerance. This study describes a novel approach for enhancing the prediction of anti-tubercular peptides by feature extraction from sequence of the peptides, selection of optimal features from the extracted features, and selection of suitable learning algorithm. Firstly, we extract different sequence features by using iFeature web server. Then, the optimal features are obtained by using a novel divergence measure-based intuitionistic fuzzy rough sets-assisted feature selection technique. Furthermore, an attempt has been made to develop models using different machine learning techniques for enhancing the prediction of anti-tubercular (or anti-mycobacterial peptides) with other antibacterial peptides (ABP) as well non-antibacterial peptides (non-ABP). Moreover, the best prediction result is obtained by vote-based classifier. Using 80:20 percentage split, the proposed method performs well, with sensitivity of 92.0%, 96.4%, specificity of 83.3%, 88.4%, overall accuracy of 87.80%, 92.90%, Mathews correlation coefficient of 0.757, 0.857, AUC of 0.922, 0.914, and g-means of 87.5%, 92.3% for anti-tubercular and ABP (primary dataset), anti-tubercular and non-ABP (secondary dataset), respectively. Finally, we have evaluated the performances of different machine learning algorithms by using the reduced training sets as produced by our proposed feature selection technique as well as already existing intuitionistic fuzzy rough set based and ensemble feature selection technique. Moreover, the performance of our proposed approach is evaluated on few benchmark and AMP datasets. From the experimental results, it can be observed that our proposed method is outperforming the previous methods.





Similar content being viewed by others
References
Ashraf M, Zaman M, Ahmed M (2019) To ameliorate classification accuracy using ensemble vote approach and base classifiers. In: Abraham A, Dutta P, Mandal JK, Bhattacharya A, Dutta S (eds) Emerging technologies in data mining and information security. Springer, Berlin, pp 321–334
Atanasov KT (1999) Intuitionistic fuzzy sets: theory and applications (Studies in Fuzziness and Soft Computing), vol 35. Physica-Verlag, Heidelberg
Atanassov KT (1986) Intuitionistic fuzzy sets. Fuzzy Sets Syst 20(1):87–96
Atanassov KT (1989) More on intuitionistic fuzzy sets. Fuzzy Sets Syst 33(1):37–45
Barnagarwala T (2014) TB hospital staff live under shadow of dreaded disease, The Indian Express. Uttar Pradesh, India: IE Online Media Services
Bhasin M, Raghava GPS (2004) Classification of nuclear receptors based on amino acid composition and dipeptide composition. J Biol Chem 279(22):23262–23266
Bhat ZS, Rather MA, Maqbool M, Lah HU, Yousuf SK, Ahmad Z (2017) Cell wall: a versatile fountain of drug targets in Mycobacterium tuberculosis. Biomed Pharmacother 95:1520–1534
Blake C, Merz C (1998) UCI repository of machine learning databases
Breiman L (1996) Bagging predictors. Mach Learn 24(2):123–140
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
Bustince H, Mohedano V (1997) About the intuitionistic fuzzy set generators. Notes Intuit Fuzzy Sets 3:21–27
Cai CZ (2003) SVM-Prot: web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acids Res 31(13):3692–3697
Cai CZ, Han LY, Ji ZL, Chen YZ (2004) Enzyme family classification by support vector machines. Proteins: Struct, Funct, Bioinf 55(1):66–76
Chakrabarty K, Gedeon T, Koczy L (2003) Intuitionistic fuzzy rough set. Wiley, Hoboken, pp 211–214
Charoenkwan P, Yana J, Schaduangrat N, Nantasenamat C, Hasan MM, Shoombuatong W (2020) iBitter-SCM: identification and characterization of bitter peptides using a scoring card method with propensity scores of dipeptides. Genomics 112(4):2813–2822. https://doi.org/10.1016/j.ygeno.2020.03.019
Chen H, Yang H (2011) One new algorithm for intuitiontistic fuzzy-rough attribute reduction. J Chin Comput Syst 32(3):506–510
Chen D, Hu Q, Yang Y (2011a) Parameterized attribute reduction with Gaussian kernel based fuzzy rough sets. Inf Sci 181(23):5169–5179
Chen Z, Chen Y-Z, Wang X-F, Wang C, Yan R-X, Zhang Z (2011b) Prediction of ubiquitination sites by using the composition of k-spaced amino acid pairs. PLoS ONE 6(7):e22930
Chen D, Kwong S, He Q, Wang H (2012a) Geometrical interpretation and applications of membership functions with fuzzy rough sets. Fuzzy Sets Syst 193:122–135
Chen D, Zhang L, Zhao S, Hu Q, Zhu P (2012b) A novel algorithm for finding reducts with fuzzy rough sets. IEEE Trans Fuzzy Syst 20(2):385–389
Chen Z, Zhou Y, Song J, Zhang Z (2013) hCKSAAP_UbSite: improved prediction of human ubiquitination sites by exploiting amino acid pattern and properties. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1834(8):1461–1467
Chen Z, Zhao P, Li F, Leier A, Marquez-Lago TT, Wang Y, Webb GI, Smith AI, Daly RJ, Chou K-C, Song J (2018) iFeature: a Python package and web server for features extraction and selection from protein and peptide sequences. Bioinformatics 34(14):2499–2502
Chou K-C (2001) Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins Struct Funct Genet 43(3):246–255
Chou KC (2004) Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 21(1):10–19
Çoker D (1998) Fuzzy rough sets are intuitionistic L-fuzzy sets. Fuzzy Sets Syst 96(3):381–383
Cornelis C, De Cock M, Kerre EE (2003) Intuitionistic fuzzy rough sets: at the crossroads of imperfect knowledge. Expert Syst 20(5):260–270
De SK, Biswas R, Roy AR (2016) Intuitionistic fuzzy database. IEEE, New York, p 43-31
Degang C, Suyun Z (2010) Local reduction of decision system with fuzzy rough set. Fuzzy Sets Syst 161(13):1871–1883
Ding C, Yuan L-F, Guo S-H, Lin H, Chen W (2012) Identification of mycobacterial membrane proteins and their types using over-represented tripeptide compositions. J Proteom 77:321–328
Dubchak I, Muchnik I, Holbrook SR, Kim SH (1995) Prediction of protein folding class using global description of amino acid sequence. Proc Natl Acad Sci 92(19):8700–8704
Dubchak I, Muchnik I, Mayor C, Dralyuk I, Kim S-H (1999) Recognition of a protein fold in the context of the SCOP classification. Struct Funct Genet 35(4):401–407
Dubois D, Prade H (1990) Rough fuzzy sets and fuzzy rough sets. Int J General Syst 17(2–3):191–209
Dubois D, Prade H (1992) Putting rough sets and fuzzy sets together. Intelligent decision support. Springer, Cham, pp 203–232
Esmail H, Maryam J, Habibolla L (2013) Rough set theory for the intuitionistic fuzzy information. Syst Int J Modern Math Sci 6(3):132–143
Feng Z-P, Zhang C-T (2000) Prediction of membrane protein types based on the hydrophobic index of amino acids. J Protein Chem 19(4):269–275
Frank E, Witten IH (1998) Generating accurate rule sets without global optimization
Grabisch M, Murofushi T, Sugeno M (2000) Fuzzy measures and integrals-theory and applications. Physica Verlag, Berlin
Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH (2009) The WEKA data mining software. ACM SIGKDD Explor Newslett 11(1):10
Han LY (2004) Prediction of RNA-binding proteins from primary sequence by a support vector machine approach. RNA 10(3):355–368
Horne DS (1988) Prediction of protein helix content from an autocorrelation analysis of sequence hydrophobicities. Biopolymers 27(3):451–477
Houben RM, Dodd PJ (2016) The global burden of latent tuberculosis infection: a re-estimation using mathematical modelling. PLoS Med 13(10):e1002152
Hu Q, Yu D, Xie Z (2006) Information-preserving hybrid data reduction based on fuzzy-rough techniques. Pattern Recogn Lett 27(5):414–423
Hu Q, Zhang L, Chen D, Pedrycz W, Yu D (2010) Gaussian kernel based fuzzy rough sets: model, uncertainty measures and applications. Int J Approx Reason 51(4):453–471
Huang B, Li HX, Wei D-K (2012) Dominance-based rough set model in intuitionistic fuzzy information systems. Knowl-Based Syst 28:115–123
Huang B, Zhuang Y-L, Li H-X, Wei D-K (2013) A dominance intuitionistic fuzzy-rough set approach and its applications. Appl Math Model 37(12–13):7128–7141
Iancu I (2014) Intuitionistic fuzzy similarity measures based on Frank t-norms family. Pattern Recogn Lett 42:128–136
Jain P, Tiwari AK, Som T (2020) A fitting model based intuitionistic fuzzy rough feature selection. Eng Appl Artif Intell 89:103421
Jena S, Ghosh S, Tripathy B (2002) Intuitionistic fuzzy rough sets. Notes on Intuitionistic Fuzzy Sets 8(1):1–18
Jensen R, Shen Q (2004a) Fuzzy–rough attribute reduction with application to web categorization. Fuzzy Sets Syst 141(3):469–485
Jensen R, Shen Q (2004b) Semantics-preserving dimensionality reduction: rough and fuzzy-rough-based approaches. IEEE Trans Knowl Data Eng 16(12):1457–1471
Jensen R, Shen Q (2005) Fuzzy-rough data reduction with ant colony optimization. Fuzzy Sets Syst 149(1):5–20
Jensen R, Shen Q (2007) Fuzzy-rough sets assisted attribute selection. IEEE Trans Fuzzy Syst 15(1):73–89
Jensen R, Shen Q (2008) Computational intelligence and feature selection: rough and fuzzy approaches. Wiley, Hoboken
Jensen R, Shen Q (2009) New approaches to fuzzy-rough feature selection. IEEE Trans Fuzzy Syst 17(4):824–838
Kalmegh S (2015) Analysis of weka data mining algorithm reptree, simple cart and randomtree for classification of indian news. Int J Innov Sci Eng Technol 2(2):438–446
Kawashima S (2000) AAindex: amino acid index database. Nucleic Acids Res 28(1):374
Kawashima S, Pokarowski P, Pokarowska M, Kolinski A, Katayama T, Kanehisa M (2007) AAindex: amino acid index database, progress report 2008. Nucleic Acids Res 36:D202–D205
Kittler J, Hatef M, Duin RPW, Matas J (1998) On combining classifiers. IEEE Trans Pattern Anal Mach Intel 20(3):226–239
Kubat M, Holte R, Matwin S (1997) Learning when negative examples abound. Springer, Berlin, pp 146–153
Kumar P, Vadakkepat P, Poh LA (2011) Fuzzy-rough discriminative feature selection and classification algorithm, with application to microarray and image datasets. Appl Soft Comput 11(4):3429–3440
Kuncheva LI (2004) Combining pattern classifiers. Wiley, Hoboken
Li L-Q, Wang X-L, Liu Z-X, Xie W-X (2019) A novel intuitionistic fuzzy clustering algorithm based on feature selection for multiple object tracking. Int J Fuzzy Syst 21:1613–1628
Lin Z, Pan X-M (2001) Accurate prediction of protein secondary structural content. J Protein Chem 20(3):217–220
Ling CX, Huang J, Zhang H (2003) AUC: a better measure than accuracy in comparing learning algorithms. Lecture notes in computer science. Springer, Berlin, pp 329–341
Lu Y-L, Lei Y-J, Hua JX (2009) Attribute reduction based on intuitionistic fuzzy rough set. Control Decis 3:003
Manavalan B, Govindaraj RG, Shin TH, Kim MO, Lee G (2018a) iBCE-EL: a new ensemble learning framework for improved linear B-cell epitope prediction. Front Immunol 9:1695
Manavalan B, Basith S, Shin TH, Wei L, Lee G (2018b) mAHTPred: a sequence-based meta-predictor for improving the prediction of anti-hypertensive peptides using effective feature representation. Bioinformatics 35(16):2757–2765
Manavalan B, Basith S, Shin TH, Wei L, Lee G (2019) AtbPpred: a robust sequence-based prediction of anti-tubercular peptides using extremely randomized trees. Comput Struct Biotechnol J 17:972–981
Montes I, Janis V, Montes S (2011) An axiomatic definition of divergence for intuitionistic fuzzy sets. In: Proceedings of the 7th conference of the European Society for Fuzzy Logic and Technology (EUSFLAT-11), pp 547–553. https://doi.org/10.2991/eusflat.2011.38
Montes I, Pal NR, Janiš V, Montes S (2015) Divergence measures for intuitionistic fuzzy sets. IEEE Trans Fuzzy Syst 23(2):444–456
Nanda S, Majumdar S (1992) Fuzzy rough sets. Fuzzy Sets Syst 45(2):157–160
Neumann U, Genze N, Heider D (2017) EFS: an ensemble feature selection tool implemented as R-package and web-application. BioData mining 10(1):1–9
Platt J (1998) Sequential minimal optimization: a fast algorithm for training support vector machines
Rizvi S, Naqvi HJ, Nadeem D (2002) Rough intuitionistic fuzzy sets. Springer, Berlin, pp 101–104
Rodriguez JJ, Kuncheva LI, Alonso CJ (2006) Rotation forest: a new classifier ensemble method. IEEE Trans Pattern Anal Mach Intel 28(10):1619–1630
Ross Quinlan J (1993) C4. 5: programs for machine learning. Mach Learn 16(3):235–240
Saha I, Maulik U, Bandyopadhyay S, Plewczynski D (2011) Fuzzy clustering of physicochemical and biochemical properties of amino acids. Amino Acids 43(2):583–594
Samanta S, Mondal T (2001) Intuitionistic fuzzy rough sets and rough intuitionistic fuzzy sets. J Fuzzy Math 9(3):561–582
Saravanan V, Gautham N (2015) Harnessing computational biology for exact linear B-cell epitope prediction: a novel amino acid composition-based feature descriptor. OMICS: A J Integr Biol 19(10):648–658
Sheeja T, Kuriakose AS (2018) A novel feature selection method using fuzzy rough sets. Comput Ind 97:111–121
Shen J, Zhang J, Luo X, Zhu W, Yu K, Chen K, Li Y, Jiang H (2007) Predicting protein-protein interactions based only on sequences information. Proc Natl Acad Sci 104(11):4337–4341
Shreevastava S, Tiwari AK, Som T (2018a) Intuitionistic fuzzy neighborhood rough set model for feature selection. Int J Fuzzy Syst Appl 7(2):75–84
Shreevastava S, Tiwari A, Som T (2018b) Feature subset selection of semi-supervised data: an intuitionistic fuzzy-rough set-based concept. Springer, Berlin, pp 303–315
Singh S, Shreevastava S, Som T, Jain P (2019) Intuitionistic fuzzy quantifier and its application in feature selection. Int J Fuzzy Syst 21(2):441–453
Sokal RR, Thomson BA (2005) Population structure inferred by local spatial autocorrelation: an example from an Amerindian tribal population. Am J Phys Anthropol 129(1):121–131
Spänig S, Heider D (2019) Encodings and models for antimicrobial peptide classification for multi-resistant pathogens. BioData Mining 12(1):7
Squeglia F, Ruggiero A, Berisio R (2018) Chemistry of peptidoglycan in Mycobacterium tuberculosis life cycle: an off-the-wall balance of synthesis and degradation. Chem—A Eur J 24(11):2533–2546
Suyun Z, Tsang E, Degang C (2009) The model of fuzzy variable precision rough sets. IEEE Trans Fuzzy Syst 17(2):451–467
Tan A, Wu W-Z, Qian Y, Liang J, Chen J, Li J (2018) Intuitionistic fuzzy rough set-based granular structures and attribute subset selection. IEEE Trans Fuzzy Syst 27(3):527–539
Teng T, Liu J, Wei H (2015) Anti-Mycobacterial Peptides: from Human to Phage. Cell Physiol Biochem 35(2):452–466
Thakur N, Qureshi A, Kumar M (2012) AVPpred: collection and prediction of highly effective antiviral peptides. Nucleic Acids Res 40(W1):W199–W204
Tiwari AK, Shreevastava S, Shukla KK, Subbiah K (2018a) New approaches to intuitionistic fuzzy-rough attribute reduction. J Intel Fuzzy Syst 34(5):3385–3394
Tiwari AK, Shreevastava S, Som T, Shukla KK (2018b) Tolerance-based intuitionistic fuzzy-rough set approach for attribute reduction. Expert Syst Appl 101:205–212
Tomii K, Kanehisa M (1996) Analysis of amino acid indices and mutation matrices for sequence comparison and structure prediction of proteins. Protein Eng Des Select 9(1):27–36
Tsang EC, Degang C, Yeung DS, Xi-Zhao W, Lee J (2008) Attributes reduction using fuzzy rough sets. IEEE Trans Fuzzy Syst 16(5):1130–1141
Usmani SS, Bhalla S, Raghava GP (2018a) Prediction of antitubercular peptides from sequence information using ensemble classifier and hybrid features. Front Pharmacol 9:954
Usmani SS, Kumar R, Kumar V, Singh S, Raghava GPS (2018) AntiTbPdb: a knowledgebase of anti-tubercular peptides. Database
Velayati AA, Farnia P, Hoffner S (2018) Drug-resistant Mycobacterium tuberculosis: epidemiology and role of morphological alterations. J Glob Antimicrob Resist 12:192–196
W. H. Organization (2016) Global tuberculosis control: WHO report 2016. Report No, WHO/HTM/TB/2016.13. Geneva, World Health Organization
W. H. Organisation (2017) Global tuberculosis report 2017, WHO Geneva, Switzerland
Wang C, Shao M, He Q, Qian Y, Qi Y (2016) Feature subset selection based on fuzzy neighborhood rough sets. Knowl-Based Syst 111:173–179
Wang J, Li J, Yang B, Xie R, Marquez-Lago TT, Leier A, Hayashida M, Akutsu T, Zhang Y, Chou K-C, Selkrig J, Zhou T, Song J, Lithgow T (2018a) Bastion3: a two-layer ensemble predictor of type III secreted effectors. Bioinformatics 35(12):2017–2028
Wang J, Yang B, Leier A, Marquez-Lago TT, Hayashida M, Rocker A, Zhang Y, Akutsu T, Chou K-C, Strugnell RA, Song J, Lithgow T (2018b) Bastion6: a bioinformatics approach for accurate prediction of type VI secreted effectors. Bioinformatics 34(15):2546–2555
Wang C, Huang Y, Shao M, Fan X (2019a) Fuzzy rough set-based attribute reduction using distance measures. Knowl-Based Syst 164:205–212
Wang C, Shi Y, Fan X, Shao M (2019b) Attribute reduction based on k-nearest neighborhood rough sets. Int J Approx Reason 106:18–31
Yager RR (1979) On the measure of fuzziness and negation part I: membership in the unit interval. Taylor & Francis, London
Yager RR (1980) On a general class of fuzzy connectives. Fuzzy Sets Syst 4(3):235–242
Yi H-C, You Z-H, Zhou X, Cheng L, Li X, Jiang T-H, Chen Z-H (2019) ACP-DL: a deep learning long short-term memory model to predict anticancer peptides using high-efficiency feature representation. Mol Ther – Nucleic Acids 17:1–9
Zhang Z (2016) Attributes reduction based on intuitionistic fuzzy rough sets. J Intel Fuzzy Syst 30(2):1127–1137
Zhang X, Zhou B, Li P (2012) A general frame for intuitionistic fuzzy rough sets. Inf Sci 216:34–49
Zhang L, Zhan J, Xu Z, Alcantud JCR (2019) Covering-based general multigranulation intuitionistic fuzzy rough sets and corresponding applications to multi-attribute group decision-making. Inf Sci 494:114–140
Acknowledgements
This research work is funded by UGC Research Fellowship, India (Grant No: 3600/(PWD)(NET-NOV2017)) awarded to first author.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by V. Loia.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jain, P., Tiwari, A.K. & Som, T. Enhanced prediction of anti-tubercular peptides from sequence information using divergence measure-based intuitionistic fuzzy-rough feature selection. Soft Comput 25, 3065–3086 (2021). https://doi.org/10.1007/s00500-020-05363-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-020-05363-z