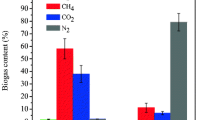
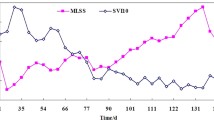
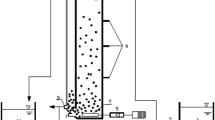
Abstract
Aerobic granular sludge degrading recalcitrant compounds are generally hard to be cultivated. This study investigated the feasibility of cultivating 2,4-dichlorophenoxyacetic acid (2,4-d) degrading aerobic granules using half-matured sludge granules pre-grown on glucose as the seeds and bioaugmentation with a 2,4-d degrading strain Achromobacter sp. QXH. Results showed that bioaugmentation promoted the steady transformation of glucose-grown granules to 2,4-d degrading sludge granules and fast establishment of 2,4-d degradation ability. The 2,4-d degradation rate of the bioaugmented granules was enhanced by 36–62 % compared to the control at 2,4-d concentrations of 144–565 mg/L on Day 18. The inoculated strain was incorporated into the half-matured granules successfully and survived till the end of operation (220 days). Sludge granules at a mean size of 420 µm and capable of utilizing 500 mg/L 2,4-d as the sole carbon source were finally obtained. Sludge microbial community shifted slightly during the whole operation and the dominant bacteria species belonged to Proteobacteria.





Similar content being viewed by others
References
Dorigo U, Bourrain X, Berard A, Leboulanger C (2004) Seasonal changes in the sensitivity of river microalgae to atrazine and isoproturon along a contamination gradient. Sci Total Environ 318:101–114
Macur RE, Wheeler JT, Burr MD, Inskeep WP (2007) Impacts of 2,4-d application on soil microbial community structure and on populations associated with 2,4-D degradation. Microbiol Res 162:37–45
Mangat SS, Elefsiniotis P (1999) Biodegradation of the herbicide 2,4-dichlorophenoxyacetic acid (2,4-D) in sequencing batch reactors. Water Res 33:861–867
Quan XC, Tang H, Xiong WC, Yang ZF (2010) Bioaugmentation of aerobic sludge granules with a plasmid donor strain for enhanced degradation of 2,4-dichlorophenoxyacetic acid. J Hazard Mater 179:1136–1142
Quan XC, Tang H, Ma JY (2011) Effects of gene augmentation on the removal of 2,4-dichlorophenoxyacetic acid in a biofilm reactor under different scales and substrate conditions. J Hazard Mater 185:689–695
Tay STL, Moy BYP, Jiang HL, Tay JH (2005) Rapid cultivation of stable aerobic phenol-degrading granules using acetate-fed granules as microbial seed. J Biotechnol 115:387–395
Wang SG, Liu XW, Zhang HY, Gong WX, Sun XW, Gao BY (2007) Aerobic granulation for 2,4-dichlorophenol biodegradation in a sequencing batch reactor. Chemosphere 69:769–775
Tay STL, Zhuang WQ, Tay JH (2005) Start-up, microbial community analysis and formation of aerobic granules in a tert-butyl alcohol degrading sequencing batch reactor. Environ Sci Technol 39:5774–5780
Yi S, Zhuang WQ, Wu B, Tay STL, Tay JH (2006) Biodegradation of p-nitrophenol by aerobic granules in a sequencing batch reactor. Environ Sci Technol 40:2396–2401
Zhu L, Xu XY, Luo WG, Tian ZJ, Lin HZ, Zhang NN (2008) A comparative study on the formation and characterization of aerobic 4-chloroaniline-degrading granules in SBR and SABR. Appl Microbiol Biot 79:867–874
Quan XC, Shi HC, Liu H, Lv PP, Qian Y (2004) Enhancement of 2,4-dichlorophenol degradation in conventional activated sludge systems bioaugmented with mixed special culture. Water Res 38:245–253
Yu FB, Ali SW, Guan LB, Li SP, Zhou S (2010) Bioaugmentation of a sequencing batch reactor with Pseudomonas putida ONBA-17, and its impact on reactor bacterial communities. J Hazard Mater 176:20–26
Quan XC, Shi HC, Wang JL, Qian Y (2003) Biodegradation of 2,4-dichlorophenol in sequencing batch reactors augmented with immobilized mixed culture. Chemosphere 50:1069–1074
van Limbergen H, Top EM, Verstraete W (1998) Bioaugmentation in activated sludge: current features and future perspectives. Appl Microbiol Biot 50:16–23
Jiang HL, Tay JH, Maszenan AM, Tay STL (2006) Enhanced phenol biodegradation and aerobic granulation by two coaggregating bacterial strains. Environ Sci Technol 40:6137–6142
Ivanov V, Wang XH, Tay STL, Tay JH (2006) Bioaugmentation and enhanced formation of microbial granules used in aerobic wastewater treatment. Appl Microbiol Biot 7:374–381
Liu YQ, Moy B, Kong YH, Tay JH (2010) Formation, physical characteristics and microbial community structure of aerobic granules in a pilot-scale sequencing batch reactor for real wastewater treatment. Enzyme Microb Technol 46:520–525
Ermakova IT, Shushkova TV, Leontevskii AA (2008) Microbial degradation of organophosphonates by soil bacteria. Microbiology 77:615–620
Tanaka T, Hachiyanagi H, Yamamoto N, Iijima T, Kido Y, Uyeda M, Takahama K (2009) Biodegradation of endocrine-disrupting chemical aniline by microorganisms. J Health Sci 55:625–630
Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jørgensen A, Molin S, Tolker-Nielsen T (2003) Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48:1511–1524
Xiong WC, Quan XC, Ma JY, Wang R (2010) Study on gfp labeling of a 2,4-D degrading strain and its detection in a wastewater biotreatment system. Environ Sci 31:1864–1870
Teske A, Wawer C, Muyzer G, Ramsing NB (1996) Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by Most-Probable-Number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl Environ Microb 62:1405–1415
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microb 59:695–700
APHA (1998) Standard methods for the examination of water and wastewater, twentieth edn. American Public Health Association/American Water Works Association/Water Environment Federation, Washington DC
Eberl L, Schulze R, Ammendola A, Geisenberger O, Erhart R, Sternberg C, Molin S, Amann R (1997) Use of green fluorescent protein as a marker for ecological studies of activated sludge communities. FEMS Microbiol Lett 149:77–83
Geisenberger O, Ammendola A, Christensen BB, Molin S, Schleifer KH, Eberl L (1999) Monitoring the conjugal transfer of plasmid RP4 in activated sludge and in situ identification of the transconjugants. FEMS Microbiol Lett 174:9–17
Jiang HL, Maszenan AM, Tay JH (2007) Bioaugmentation and coexistence of two functionally similar bacterial strains in aerobic granules. Appl. Microbiol Biot 75:1191–1200
Wilderer PA, Rubio MA, Davids L (1991) Impact of the addition of pure cultures on the performance of mixed culture reactors. Water Res 25:1307–1313
Fulthorpe RR, Mcgowan C, Maltseva OV, Holben WE, Tiedje JM (1995) 2,4-dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl Environ Microb 61:3274–3281
Kamagata Y, Fulthorpe RR, Tamura K, Takami H, Forney LJ, Tiedje JM (1997) Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl Environ Microb 63:2266–2272
McGowan C, Fulthorpe R, Wright A, Tiedje JM (1998) Evidence for interspecies gene transfer in the evolution of 2,4-dichlorophenoxyacetic acid degraders. Appl Environ Microb 64:4089–4092
Cavalca L, Hartmann A, Rouard N, Soulas G (1999) Diversity of tfdC genes: distribution and polymorphism among 2,4-dichlorophenoxyacetic acid degrading soil bacteria. FEMS Microbiol Ecol 29:45–58
Vallaeys T, Courde L, Mc Gowan C, Wright AD, Fulthorpe RR (1999) Phylogenetic analyses indicate independent recruitment of diverse gene cassettes during assemblage of the 2,4-D catabolic pathway. FEMS Microbiol Ecol 28:373–382
Zhang J, Zheng JW, Hang BJ, Ni YY, He JA, Li SP (2011) Rhodanobacter xiangquanii sp. nov., a novel anilofos-degrading bacterium isolated from a wastewater treating system. Curr Microbiol 62:645–649
Zitomer DH, Duran M, Albert R, Guven E (2007) Thermophilic aerobic granular biomass for enhanced settleability. Water Res 41:819–825
Toyama T, Sato Y, Inoue D, Sei K, Chang YC, Kikuchi S, Ike M (2009) Biodegradation of bisphenol A and bisphenol F in the rhizosphere sediment of Phragmites australis. J Biosci Bioeng 108:147–150
Huang HD, Wang W, Ma T, Li GQ, Liang FL, Liu RL (2009) Sphingomonas sanxanigenens sp. nov., isolated from soil. Int J Syst Evol Micr 59:719–723
Hashimoto T, Onda K, Morita T, Luxmy BS, Tada K, Miya A, Murakami T (2010) Contribution of the estrogen-degrading bacterium Novosphingobium sp. Strain JEM-1 to estrogen removal in wastewater treatment. J Environ Eng ASCE 136:890–896
Liang QF, Lloyd-Jones G (2010) Sphingobium scionense sp. nov., an aromatic hydrocarbon-degrading bacterium isolated from contaminated sawmill soil. Int J Syst Evol Micr 60:413–416
Sipila TP, Vaisanen P, Paulin L, Yrjala K (2010) Sphingobium sp. HV3 degrades both herbicides and polyaromatic hydrocarbons using ortho- and meta-pathways with differential expression shown by RT-PCR. Biodegradation 21:771–784
Acknowledgments
This research was supported by “National Natural Science Foundation of China” (No. 51178049). The authors wish to express their gratitude to Prof. Søren Molin in Technical University of Denmark for the donation some strains required for gfp labeling.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Quan, X., Ma, J., Xiong, W. et al. Bioaugmentation of half-matured granular sludge with special microbial culture promoted establishment of 2,4-dichlorophenoxyacetic acid degrading aerobic granules. Bioprocess Biosyst Eng 38, 1081–1090 (2015). https://doi.org/10.1007/s00449-014-1350-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-014-1350-y