Abstract
Podocytes are specialized epithelial cells used for glomerular filtration in the kidney. They can be divided into the cell body, primary process and foot process. Here, we describe two useful methods for the three-dimensional(3D) visualization of these subcellular compartments in rodent podocytes. The first method, field-emission scanning electron microscopy (FE-SEM) with conductive staining, is used to visualize the luminal surface of numerous podocytes simultaneously. The second method, focused-ion beam SEM (FIB-SEM) tomography, allows the user to obtain serial images from different depths of field, or Z-stacks, of the glomerulus. This allows for the 3D reconstruction of podocyte ultrastructure, which can be viewed from all angles, from a single image set. This is not possible with conventional FE-SEM. The different advantages and disadvantages of FE-SEM and FIB-SEM tomography compensate for the weaknesses of the other. The combination renders a powerful approach for the 3D analysis of podocyte ultrastructure. As a result, we were able to identify a new subcellular compartment of podocytes, “ridge-like prominences” (RLPs).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Podocytes are specialized epithelial cells used for glomerular filtration, which are classically divided into three subcellular compartments: the cell body, the primary process and the foot process. Here, we describe two useful methods for the three-dimensional(3D) visualization of these subcellular compartments in rodent podocytes. The first method, field-emission scanning electron microscopy (FE-SEM), is used to visualize the luminal surface of numerous podocytes simultaneously (Kinugasa et al. 2011; Masum et al. 2018; Tagawa et al. 2016). The second method, focused-ion beam SEM (FIB-SEM) tomography, allows the user to obtain serial images from different depths of field, or Z-stacks, of the glomerulus (Arkill et al. 2014; Burghardt et al. 2015). This allows for the 3D reconstruction of podocyte ultrastructure, which can be viewed from all angles, in a single image set. This is not possible with conventional FE-SEM. Our research group analyzed the 3D ultrastructure of podocytes in health, disease and development by utilizing FE-SEM and FIB-SEM tomography (Ichimura et al. 2017; Ichimura et al. 2019; Ichimura et al. 2015; Ichimura and Sakai 2017; Kawasaki et al. 2019). Here, we demonstrate their advantages and limitations in podocyte ultrastructural study.
Materials and methods
Animals
For investigating the 3D ultrastructure of normal podocytes, we used 10-week-old male rats (Charles River Japan, Yokohama, Japan). To induce puromycin aminonucleoside (PAN) nephrosis, we intraperitoneally administered PAN (15 mg/100 g body weight; Sigma-Aldrich, St. Louis, MO, USA) to 6-week-old male Wistar rats (Charles River Japan). All procedures performed on the laboratory animals were approved by the Institutional Animal Care and Use Committee of Juntendo University School of Medicine (Approval no. 300226).
Perfusion fixation of kidneys
An arterial catheter (PE50, BD Intramedic Polyethylene Tubing, Thermo Fisher Scientific) was retrogradely inserted via the abdominal aorta, below the renal arteries, under anaesthesia with pentobarbital. To wash out blood from the vascular system, the inferior vena cava was incised, then, physiological saline was perfused for 2–3 min. The abdominal organs, including kidneys, whiten due to blood removal. Subsequently, 2.5% glutaraldehyde/0.1 M phosphate buffer (PB) was perfused for 3 min, which makes the abdominal organs become hard and turn yellowish. Perfusion of saline and fixative should be performed at 180–200 mmHg using a constant-pressure perfusion apparatus, such as a VPF-1 perfusion apparatus (Nisshin EM, Tokyo, Japan). The fixed kidneys were immersed in the same fixative for 2–3 days at 4 °C.
FE-SEM with conductive staining
For the step-by-step protocol of FE-SEM with conductive staining, see Supplementary Method S1.
Conductive staining
The cortex of the fixed kidney was cut into small cubes (approximately 4 × 4 × 2 mm). After being washed with 0.1 M PB, the cubes were processed with conductive staining. First, the cubes were immersed in 1% osmium tetroxide (OsO4) in 0.1 M PB for 30 min at 24 °C; washed with 0.1 M PB for 5 min three times; and then immersed in 1% low molecular weight tannic acid (LMW-TA, Electron Microscopy Sciences, Hatfield, PA, USA) in distilled water (DW), for 2 h at RT. After the cubes were washed with DW for 5 min three times, the same staining was repeated twice. However, the OsO4 was diluted with DW.
Dehydration, drying and mounting
The stained samples were dehydrated with graded series of ethanol (50% for 5 min; 70% for 5 min; 90% for 5 min; 100% for 5 min, five times) and then immersed in t-butyl alcohol for 5 min, three times. The samples were freeze-dried with an ES-2030 freeze dryer (Hitachi High-Technologies, Tokyo, Japan). The dried specimens were mounted on aluminum stubs with carbon tape (Cat #732, Nissin EM) and paste (Pelco Colloidal Graphite, Ted Pella, Inc. Redding, CA, USA). The mounted specimens were coated with osmium with an OPC80T osmium plasma coater (Filgen, Inc., Nagoya, Japan).
FE-SEM
The samples were observed with an S-4800 FE-SEM (Hitachi High-Technologies). Regions of interest were imaged using a backscattered electron detector with an acceleration voltage of 3 kV. The pixel dimensions for a recorded image were 3072 × 2048 pixels. Individual podocytes are colored with different transparent colors using Adobe Photoshop or Illustrator. This procedure was made easier by using a Cintiq 27QHD interactive pen display (Wacom, Tokyo, Japan).
FIB-SEM tomography and 3D reconstruction
For the step-by-step protocol of FIB-SEM tomography and 3D reconstruction, see Supplementary Method S2.
Combinatorial heavy metal block staining
The cortex of the fixed kidney was cut into 250-μm-thick slices with a DTK-1000 Microslicer (Dosaka EM, Kyoto, Japan). The slices were processed using the combinatorial heavy metal staining protocol. This protocol was designed to enhance signal for backscatter electron imaging of epoxy resin–embedded mammalian tissues, at low accelerating voltages. The slices were successively immersed in 1% OsO4, containing 1.5% potassium ferrocyanide (K4Fe(CN)6·3H2O), in 0.1 M cacodylate buffer (CB) for 1 h on ice; 1% LMW-TA (Electron Microscopy Sciences) in 0.1 M CB, for 4 h at 24 °C; 2% aqueous OsO4 for 30 min at 24 °C; and 1% aqueous uranyl acetate (UO2(CH3COO)2·2H2O) overnight at 24 °C. Between each staining step, the slices were washed with 0.1 M CB or DW for 5 min, three times.
Dehydration and embedding
The stained samples were then dehydrated with a graded series of ethanol (50% for 5 min; 70% for 5 min; 90% for 5 min; 100% for 5 min, five times). The dehydrated samples were successively immersed in propylene oxide for 5 min, twice; 50% epoxy resin in propylene oxide, on a rotational mixer, for 3 h; and then pure epoxy resin, on a rotational mixer, overnight. Subsequently, the samples were embedded in newly made pure epoxy resin and hardened in a heated air incubator for three to 5 days. We used a hardening formula of Oken Epok 812 epoxy resin kit (Oken-shoji, Tokyo, Japan).
Mounting and coating
The surface of resin-embedded tissues was exposed using a diamond knife on an Ultracut UCT (Leica Biosystems). The block was mounted onto an HV-8 aluminum stub (Micro Star, Tokyo, Japan). The space between the block and stub was filled with carbon paste (Pelco Colloidal Graphite, Ted Pella, Inc.) and then coated with a thin layer of platinum-palladium, using an MC1000 ion sputter coater (Hitachi High-Technologies) to prevent charging.
FIB-SEM tomography
The exposed surface of embedded tissue was imaged with a Helios NanoLab 660 FIB-SEM (Thermo Fisher Scientific, Waltham, MA, USA), at a high acceleration voltage of 10 kV, to find the area of interest. To prevent beam damage, a platinum layer was deposited on the area of interest, with a 2.5-nA beam current, where gallium ions were accelerated by a voltage of 30 kV. To make the new block-face (imaging-face), a trench was cut using FIB-milling, with a 25-nA beam current, where gallium ions were accelerated by a voltage of 30 kV. Serial FIB-SEM images were acquired with Auto Slice and View imaging software (Thermo Fisher Scientific) on a Helios NanoLab 660 FIB-SEM. New surfaces for serial block-face imaging were generated using FIB-milling, with a 0.77-nA beam current. Serial block-face images (300–800) were obtained every 50 nm in depth, with a backscattered electron detector at an acceleration voltage of 2.0 kV. The pixel size of each FIB-SEM image was 13.5 × 17.1 × 50 nm/pixel (width × height × depth). The pixel dimensions for a recorded image were 3072 × 2048 pixels. Thus, the dimension of the serial images acquired using FIB-SEM was 41.5 × 35.0 × 50 μm (width × height × depth).
3D reconstruction
The alignment of serial FIB-SEM images, segmentation of target podocytes and 3D reconstruction were performed using AMIRA 6.1 reconstruction software (Thermo Fisher Scientific). We also used a Cintiq 27QHD interactive pen display for the segmentation procedure (Wacom). The 3D images could be presented as a movie or 3D-printed model.
Results and discussion
FE-SEM
The FE-SEM procedure, for 3D ultrastructural analysis of podocytes, is divided into eight major steps (Fig. 1): (1) perfusion fixation at a high hydrostatic pressure; (2) conductive staining; (3) dehydration; (4)freeze-drying;(5) mounting on a stub; (6) coating with heavy metal; (7) image acquisition; (8) coloring (optional) (for the step-by-step protocol for FE-SEM, see Supplementary Method S1). All steps should be adequately performed to prevent artificial structural alterations. In particular, the first and second steps are crucial in realizing a clear presentation of the podocyte surface ultrastructure.
Outline protocol for FE-SEM. Procedure for FE-SEM is divided into eight steps (1–8). (a) Freeze-dried samples on an aluminum stub. (b) FE-SEM image of normal rat podocytes. (b′) Individual podocytes in the FE-SEM image, which have been colored by Adobe Illustrator image software. The step-by-step protocols for FE-SEM are described in Supplementary Method S1.
It is essential that glomerular capillaries are fixed in the distended state (using step 1), to prevent their collapse and, thus, distortion of the podocyte architecture.
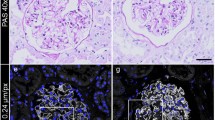
The staining procedure is often conducted only using 1% OsO4. This is a simple method but the podocytes can end up appearing distorted and somewhat shrunken (Fig. 2b). Instead, we consider the more invested conductive staining method, which maintains the size of the cells and the integrity of the glomerular capillaries without artificial distortions during subsequent steps (Fig. 2a).
Efficacy of conductive staining for preservation of podocyte ultrastructure in FE-SEM. (a, a′) Glomerular capillaries, which were fixed in the distended state by perfusion fixation, maintain distended during freeze-drying due to conductive staining. (a′) The surface of cell body, primary processes and foot processes also remained smooth, without shrinkage, due to the use of conductive staining. (b, b′) Simple OsO4 staining was incapable of preventing the collapse of glomerular capillaries and the shrinkage of podocytes. (a, b) Low-magnification images of a glomerulus in a normal 10-week-old Wistar rat. (a′, b′) High-magnification images showing the three compartments of podocytes. Scale bars: 50 μm in (a, b); 10 μm in (a′, b′).
The coloring of individual podocytes, in the FE-SEM images, is useful for understanding the architecture and mutual relationship of the podocytes (Figs. 1b′ and 3). Such coloring can be easily performed by commercially available image software, such as Adobe Photoshop or Illustrator.
FE-SEM images showing the 3D structural alteration of podocytes during PAN nephrosis development. Individual podocytes are shown in different colors. (a) Healthy (control) podocytes. The cell body projected primary processes, which protruded numerous fine foot processes that interdigitated with those of the neighboring podocytes. (b, c) Early phase (days 2–3): podocytes still maintain three subcellular compartments, although some of the primary processes have become irregularly swollen. (d) Overt proteinuria phase (day 8): podocytes are largely reorganized into a broad flattened shape—the three subcellular compartments, as found in the normal and early phase podocytes—are almost completely lost. CB, cell body; P, primary process. Scale bars: 5 μm.
FE-SEM, with conductive staining, is an easy and efficient method for visualizing the luminal surface of numerous podocytes in different glomeruli, simultaneously. It is also useful for analyzing the outline of the 3D podocyte architecture without structural distortions. However, there are several limitations. The basal surface and the regions between glomerular capillaries are quite difficult to observe by FE-SEM. Thus, it cannot be used to image the entire surface of individual podocytes.
FIB-SEM tomography
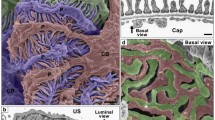
The procedure for FIB-SEM tomography, for the 3D structural analysis of podocytes, is divided into 11 major steps (Fig. 4): (1) perfusion fixation at a high hydrostatic pressure; (2) combinatorial heavy metal staining; (3) dehydration; (4) embedding in resin; (5) surface exposure; (6) mounting on a stub; (7) coating with heavy metal; (8) generating a new block-face;(9) serial image acquisition; (10) segmentation of podocytes; (11) 3D reconstruction (for the step-by-step protocol for FIB-SEM tomography, see Supplementary Method S2). All steps should be performed to prevent artificial structural alterations. In particular, steps (1), (2) and (9) are necessary to achieve a clear presentation of the podocyte ultrastructure and 3D reconstruction.
Outline protocol for FIB-SEM tomography. Procedure of FIB-SEM is divided into eleven major steps (1–11). (a) Resin-embedded block mounted on an aluminum stub and coated with platinum-palladium (Pt-Pd). (b) Schematic drawing of the positional relationships of the sample (yellow), the FIB column (pink) and the SEM column (green). The cross-sectional surface, called the “block-face” (asterisk), is generated by FIB milling. The block-face is perpendicular to the surface of the sample. The SEM column is situated obliquely to scan the block-face. (b′) Low-magnification image of the block-face. (c) The contrast-inverted serial FIB-SEM images are similar to a conventional transmission electron microscope (TEM) image of an ultrathin section. The generation and imaging of the block-face is automatically performed to acquire serial FIB-SEM images. (d) Reconstructed images of a single podocyte produced from the serial FIB-SEM images, using reconstruction software. (d′) 3D-printed podocyte. The detail and step-by-step protocols for FIB-SEM are described in Supplementary Method S2.
Combinatorial heavy metal staining is used to increase the contrast in FIB-SEM images. Several protocols for this staining method have been reported by different research groups (summarized in Kubota et al. 2018). Our protocol is relatively simple. Uranyl acetate (UO2(CH3COO)2) is generally used for this staining method; however, it is an internationally controlled material and requires extreme handling caution. Instead, samarium triacetate (Sm(CH3COO)3) or gadolinium triacetate (Gd(CH3COO)3) could be used (Nakakoshi et al. 2011; Odriozola et al. 2017).
The manual segmentation of podocytes from serial FIB-SEM images is made much simpler by using an interactive pen display (Fig. 5), which is also useful for the coloring of podocytes in FE-SEM images.
The 3D reconstruction data can be represented by a 3D-printed plastic model (Fig. 2d′) and as a movie (Supplementary Movies S1, S2 and S3). Such models and movies are useful for comprehensively demonstrating the 3D ultrastructure of podocytes, especially in histology and pathology education for medical students.
The 3D-reconstructed images of single podocytes, generated from serial FIB-SEM images, allow the visualization of the entire surface of podocytes from any angle and without interference by neighboring podocytes or the glomerular basement membrane. By observing the basal surface of single reconstructed podocytes, we found “ridge-like prominences (RLPs)”, a new subcellular compartment of podocytes, by which the foot processes are linked to the cell body and primary processes (Fig. 6)(Ichimura et al. 2017; Ichimura et al. 2015).
Structural hierarchy of podocyte subcellular compartments revealed by FIB-SEM tomography. (a) Previous simple hierarchy. The three subcellular compartments of the podocyte—cell body, primary processes and foot processes—are related in this order. (a′) New hierarchy based on our previous 3D analysis. The ridge-like prominences, which protrude from the basal surface of the cell body and primary processes, serve as an adhesion apparatus for the attachment of the cell body and primary processes to the GBM and to connect foot processes to the cell body and primary processes. (b, b′) Schematic representation of podocyte subcellular compartments based on the new hierarchy. (b′) The ridge-like prominences are colored yellow. (c, c′) 3D-reconstructed podocyte from serial FIB-SEM images. The luminal and basal surfaces of the same reconstructed podocyte are shown. CB, cell body; FP, foot process; PP, primary process; RLP, ridge-like prominence. Scale bars: 1 μm.
The multicolored reconstructed images of podocytes are useful for analyzing the relationships between these cells (Fig. 7). For instance, we can show that podocytes frequently form cytoplasmic fragmentation (Fig. 7c′) and autocellular tight junctions during the developmental period of the foot process effacement in puromycin aminonucleoside (PAN) nephrosis (Ichimura et al. 2019).
Structural alterations in basal surfaces of podocytes during PAN nephrosis development revealed by FIB-SEM tomography. The basal view of the reconstructed podocytes, which are individually shown in different colors, used to analyze structural alterations in the foot processes. (a) In healthy (control) podocytes, foot processes exhibit a uniform width. (b) In the early phase (day 4) of PAN nephrosis, podocytes lose uniformity in the foot processes. (c) Some of the regions form a large adhesive surface in this phase. (d) In the overt proteinuria phase (day 8), normal patterns of the interdigitating foot processes largely disappear and most podocytes form a large adhesive surface. (c, d) The yellow masses represent the cytoplasmic fragments of the podocyte, which do not continue to the neighboring podocytes. The cytoplasmic fragments vary in size and shape and are frequently found in PAN nephrosis. Scale bar: 2 μm.
FIB-SEM tomography is also suitable for 3D structural visualization of other cells that are entirely surrounded by the extracellular matrix. For example, mesangial cells and nephrocytes—one of the podocyte-related cells found in arthropods—includingDrosophila melanogaster(Ichimura and Sakai 2017; Kawasaki et al. 2019).
Although FIB-SEM tomography is a powerful tool for precisely analyzing the 3D ultrastructure of podocytes, this method has several technical disadvantages. The imaged region is completely lost due to the required FIB milling. Moreover, the observational area is restricted in FIB-SEM tomography, rendering it impossible to visualize the entire glomerular surface. Array tomography is a potential solution to this problem. It has been used to obtain serial images from a series of ultrathin sections mounted on silicon wafer and glass plate substrates (Koga et al. 2018; Koike et al. 2017). These ultrathin sections have been shown to be physically stable and repeatable. In future investigations, we aim to establish a “serial ultrathin sections library” of renal biopsy specimens and to carry out analysis using array tomography.
In our laboratory, we manually segmented podocytes on 200–500 serial FIB/SEM images using Amira software. This manual segmentation procedure is the main rate-determining step in our 3D structural analysis of podocytes, although it becomes easy to perform using the interactive pen display, as described above. For instance, it took approximately 25 and 50 h to segment podocytes for producing the reconstruction images shown in Figs. (6c, 7a), respectively. Various image analysis software containing an automated segmentation tool have been reported and some of them are commercially and freely available (Moen et al. 2019). However, there has been no successful case that podocytes could be automatically segmented at a high level of quality—the special complexity of podocyte structure makes it quite difficult to automatically segment them. To make the 3D ultrastructural analysis of podocytes more efficient, a novel tool needs to be developed using artificial intelligence that is specified for automated segmentation of podocytes.
Conclusion
The different advantages and disadvantages of FE-SEM and FIB-SEM tomography allow each technique to compensate for the weaknesses of the other. The combination of these produces a powerful approach for the 3D analysis of podocyte ultrastructure. As a result, we were able to identify a new subcellular compartment of podocytes, RLPs.
References
Arkill KP, Qvortrup K, Starborg T et al (2014) Resolution of the three dimensional structure of components of the glomerular filtration barrier. BMC Nephrol 15:24
Burghardt T, Hochapfel F, Salecker B et al (2015) Advanced electron microscopic techniques provide a deeper insight into the peculiar features of podocytes. Am J Physiol Renal Physiol 309:F1082–F1089
Ichimura K, Sakai T (2017) Evolutionary morphology of podocytes and primary urine-producing apparatus. Anat Sci Int 92:161–172
Ichimura K, Miyazaki N, Sadayama S et al (2015)Three-dimensional architecture of podocytes revealed by block-face scanning electron microscopy. Sci Rep 5:8993
Ichimura K, Kakuta S, Kawasaki Y et al (2017) Morphological process of podocyte development revealed by block-face scanning electron microscopy. J Cell Sci 130:132–142
Ichimura K, Miyaki T, Kawasaki Y, Kinoshita M, Kakuta S, Sakai T (2019) Morphological processes of foot process effacement in puromycin aminonucleoside nephrosis revealed by FIB/SEM tomography. J Am Soc Nephrol 30:96–108
Kawasaki Y, Matsumoto A, Miyaki T et al (2019)Three-dimensional architecture of pericardial nephrocytes in Drosophila melanogaster revealed by FIB/SEM tomography. Cell Tissue Res. https://doi.org/10.1007/s00441-00019-03037-00443
Kinugasa S, Tojo A, Sakai T et al (2011) Selective albuminuria via podocyte albumin transport in puromycin nephrotic rats is attenuated by an inhibitor of NADPH oxidase. Kidney Int 80:1328–1338
Koga D, Kusumi S, Watanabe T (2018) Backscattered electron imaging of resin-embedded sections. Microscopy (Oxf) 67:196–206
Koike T, Kataoka Y, Maeda M et al (2017) A device for ribbon collection for array tomography with scanning electron microscopy. Acta Histochem Cytochem 50:135–140
Kubota Y, Sohn J, Hatada S et al (2018) A carbon nanotube tape for serial-section electron microscopy of brain ultrastructure. Nat Commun 9:437
Masum MA, Ichii O, Elewa YHA et al (2018) Modified scanning electron microscopy reveals pathological crosstalk between endothelial cells and podocytes in a murine model of membranoproliferative glomerulonephritis. Sci Rep 8:10276
Moen E, Bannon D, Kudo T, Graf W, Covert M, Van Valen D (2019) Deep learning for cellular image analysis. Nat Methods. https://doi.org/10.1038/s41592-41019-40403-41591
Nakakoshi M, Nishioka H, Katayama E (2011) New versatile staining reagents for biological transmission electron microscopy that substitute for uranyl acetate. J Electron Microsc (Tokyo) 60:401–407
Odriozola A, Llodrá J, Radecke J, et al. (2017) High contrast staining for serial block face scanning electron microscopy without uranyl acetate. bioRxiv https://doi.org/10.1101/207472
Tagawa A, Yasuda M, Kume S et al (2016) Impaired podocyte autophagy exacerbates proteinuria in diabetic nephropathy. Diabetes 65:755–767
Acknowledgments
The authors wish to thank Mr. Takanobu Ishimura (Maxnet Co., Ltd., Tokyo, Japan) for giving a technical lecture on the reconstruction software and Dr. Kota Kato (Juntendo University) for the 3D printing.
Funding
This work was financially supported in part by the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant nos. JP15K18960, JP17K08521 to KI) and the Foundation of Strategic Research Projects in Private Universities from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) (Grant nos. S1311011, S1101009 to Juntendo University).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Not applicable.
Ethical approval
Animal experiments in this study were approved by the Institutional Animal Care and Use Committee of Juntendo University School of Medicine (approval no. 300226) and were carried out in accordance with the Guidelines for Animal Experimentation of Juntendo University School of Medicine. This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
Supplementary Method S1. Step-by-step protocol for FE-SEM. Supplementary Method S2. Step-by-step protocol for FIB-SEM tomography (DOCX 37 kb)
ESM 2
Two interdigitating podocytes in normal adult rat (MPG 71177 kb)
ESM 3.
Primary and foot processes from single podocyte in normal adult rat (MPG 72950 kb)
ESM 4.
PAN nephrotic podocytes (MPG 56523 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Miyaki, T., Kawasaki, Y., Hosoyamada, Y. et al. Three-dimensional imaging of podocyte ultrastructure using FE-SEM and FIB-SEM tomography. Cell Tissue Res 379, 245–254 (2020). https://doi.org/10.1007/s00441-019-03118-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-019-03118-3