Abstract
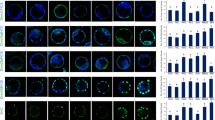
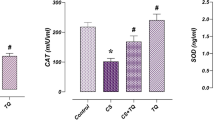
Sulforaphane (SFN) has been considered as an indirect antioxidant and potential inducer of the Nrf2-ARE pathway. This study was conducted to investigate the protective role of SFN against oxidative stress in bovine granulosa cells (GCs). GCs were collected from antral follicles (4–8 mm) and cultured according to the experimental design where group 1 = control, group 2 = treated with SFN, group 3 = treated with hydrogen peroxide (H2O2), group 4 = pretreated with SFN and then with H2O2 (protective) and group 5 = treated with H2O2 followed by SFN treatment (rescuing). Results showed that SFN pretreatment significantly increases cell viability and reduces cytotoxicity in GCs under oxidative stress. Following H2O2 exposure, expression of NRF2 was found to be significantly increased (p < 0.05) in SFN-pretreated cells, while no significant differences were observed between group 3 and group 5, although the expression was significantly increased compared to the control group. Moreover, the relative abundance of the NRF2 downstream target antioxidant genes (CAT, PRDX1, SOD1 and TXN1) were higher (fold change ranged from 7 to 14, p < 0.05) in sulforaphane pretreated GCs. Low level of ROS and lipid accumulation and higher mitochondrial activity were observed in GCs pretreated with SFN, whereas no such changes were observed in GCs treated with SFN after exposure to oxidative stress (group 5). Thus, we suggest that SFN pretreatment effectively protects GCs against oxidative damage through the activation of the NRF2-ARE pathway, whereas addition of SFN during oxidative insult failed to rescue GCs.






Similar content being viewed by others
Change history
06 November 2018
There is an error in the original publication of this paper. Figures 1-6 were shown in the wrong version, thus corrected figures provided were in this article.
References
Agarwal A, Gupta S, Sharma RK (2005) Role of oxidative stress in female reproduction. Reprod Biol Endocrinol 3(28). https://doi.org/10.1186/1477-7827-3-28
Agarwal A, Aponte-Mellado A, Premkumar BJ et al (2012) The effects of oxidative stress on female reproduction: a review. Reprod Biol Endocrinol 10:49. https://doi.org/10.1186/1477-7827-10-49
Amin A, Gad A, Salilew-Wondim D et al (2014) Bovine embryo survival under oxidative-stress conditions is associated with activity of the NRF2-mediated oxidative-stress-response pathway. Mol Reprod Dev 81:497–513
Buckley BJ, Marshall ZM, Whorton A (2003) Nitric oxide stimulates Nrf2 nuclear translocation in vascular endothelium. Biochem Biophys Res Commun 307:973–979. https://doi.org/10.1016/S0006-291X(03)01308-1
Chaudhuri D, Orsulic S, Ashok BT (2007) Antiproliferative activity of sulforaphane in Akt-overexpressing ovarian cancer cells. Mol Cancer Ther 6:334–345. https://doi.org/10.1158/1535-7163.MCT-06-0404
Chuang LT, Moqattash ST, Gretz HF et al (2007) Sulforaphane induces growth arrest and apoptosis in human ovarian cancer cells. Acta Obstet Gynecol Scand 86:1263–1268. https://doi.org/10.1080/00016340701552459
Dong Z, Shang H, Chen YQ et al (2016) Sulforaphane protects pancreatic acinar cell injury by modulating Nrf2-mediated oxidative stress and NLRP3 inflammatory pathway. Oxidative Med Cell Longev 2016:1–12. https://doi.org/10.1155/2016/7864150
Dou T, Yan M, Wang X et al (2016) Nrf2/ARE pathway involved in oxidative stress induced by paraquat in human neural progenitor cells. Oxidative Med Cell Longev 2016:1–8. https://doi.org/10.1155/2016/8923860
Dröse S, Brandt U (2012) Molecular mechanisms of superoxide production by the mitochondrial respiratory chain. Adv Exp Med Biol 748:145–169. https://doi.org/10.1007/978-1-4614-3573-0_6
Duffy DM, Stouffer RL (2003) Luteinizing hormone acts directly at granulosa cells to stimulate periovulatory processes: modulation of luteinizing hormone effects by prostaglandins. Endocrine 22:249–256. https://doi.org/10.1385/ENDO:22:3:249
Frisard M, Ravussin E (2006) Energy metabolism and oxidative stress: impact on the metabolic syndrome and the aging process. Endocrine 29:27–32. https://doi.org/10.1385/ENDO:29:1:27
Fuentes F, Paredes-Gonzalez X, Kong A-NT (2015) Dietary glucosinolates sulforaphane, phenethyl isothiocyanate, indole-3-carbinol/3,3′-diindolylmethane: anti-oxidative stress/inflammation, Nrf2, epigenetics/epigenomics and in vivo cancer chemopreventive efficacy. Curr Pharmacol Rep 1:179–196. https://doi.org/10.1007/s40495-015-0017-y
Gamet-Payrastre L, Li P, Lumeau S et al (2000) Sulforaphane, a naturally occurring isothiocyanate, induces cell cycle arrest and apoptosis in HT29 human colon cancer cells. Cancer Res 60:1426–1433. https://doi.org/10.1158/0008-5472.can-06-0139
Gorrini C, Harris IS, Mak TW (2013) Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov 12:931–947. https://doi.org/10.1038/nrd4002
Guerrero-Beltrán CE, Calderón-Oliver M, Pedraza-Chaverri J, Chirino YI (2012) Protective effect of sulforaphane against oxidative stress: recent advances. Exp Toxicol Pathol 64:503–508. https://doi.org/10.1016/j.etp.2010.11.005
He C, Li B, Song W et al (2014) Sulforaphane attenuates homocysteine-induced endoplasmic reticulum stress through Nrf-2-driven enzymes in immortalized human hepatocytes. J Agric Food Chem 62:7477–7485. https://doi.org/10.1021/jf501944u
Hemachandra LPMP, Chandrasekaran A, Melendez JA, Hempel N (2016) Regulation of the cellular redox environment by superoxide dismutases, catalase, and glutathione peroxidases during tumor metastasis. Springer International Publishing, New York, pp 51–79
Hong F, Freeman ML, Liebler DC (2005) Identification of sensor cysteines in human Keap1 modified by the cancer chemopreventive agent sulforaphane. Chem Res Toxicol 18:1917–1926. https://doi.org/10.1021/tx0502138
Itoh K, Chiba T, Takahashi S et al (1997) An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236:313–322. https://doi.org/10.1006/bbrc.1997.6943
Janssen-Heininger YMW, Mossman BT, Heintz NH et al (2008) Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic Biol Med 45:1–17. https://doi.org/10.1016/j.freeradbiomed.2008.03.011
Jin X, Wang K, Liu H et al (2016) Protection of bovine mammary epithelial cells from hydrogen peroxide-induced oxidative cell damage by resveratrol. Oxidative Med Cell Longev 2016:1–15. https://doi.org/10.1155/2016/2572175
Juengel E, Maxeiner S, Rutz J et al (2016) Sulforaphane inhibits proliferation and invasive activity of everolimus-resistant kidney cancer cells in vitro. Oncotarget 7:85208–85219. https://doi.org/10.18632/oncotarget.13421
Kim BG, Fujita T, Stankovic KM et al (2016) Sulforaphane, a natural component of broccoli, inhibits vestibular schwannoma growth in vitro and in vivo. Sci Rep 6:36215. https://doi.org/10.1038/srep36215
Klaunig JE, Kamendulis LM, Hocevar BA (2010) Oxidative stress and oxidative damage in carcinogenesis. Toxicol Pathol 38:96–109. https://doi.org/10.1177/0192623309356453
Kwak M-K, Itoh K, Yamamoto M, Kensler TW (2002) Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol Cell Biol 22:2883–2892. https://doi.org/10.1128/mcb.22.9.2883-2892.2002
Lee J, Homma T, Kurahashi T, et al (2015) Oxidative stress triggers lipid droplet accumulation in primary cultured hepatocytes by activating fatty acid synthesis. Biochem Biophys Res Commun 464:229–235. https://doi.org/10.1016/j.bbrc.2015.06.121
Li B, Kim DS, Yadav RK et al (2015) Sulforaphane prevents doxorubicin-induced oxidative stress and cell death in rat H9c2 cells. Int J Mol Med 36:53–64. https://doi.org/10.3892/ijmm.2015.2199
Liu H, Smith AJO, Lott MC et al (2013) Sulforaphane can protect Lens cells against oxidative stress: implications for cataract prevention. Invest Opthalmol Vis Sci 54:5236. https://doi.org/10.1167/iovs.13-11664
Ma Q (2013) Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol 53:401–426. https://doi.org/10.1146/annurev-pharmtox-011112-140320
Medina S, Domínguez-Perles R, Moreno DA et al (2015) The intake of broccoli sprouts modulates the inflammatory and vascular prostanoids but not the oxidative stress-related isoprostanes in healthy humans. Food Chem 173:1187–1194. https://doi.org/10.1016/J.FOODCHEM.2014.10.152
Nair S, Li W, Kong A-NT (2007) Natural dietary anti-cancer chemopreventive compounds: redox-mediated differential signaling mechanisms in cytoprotection of normal cells versus cytotoxicity in tumor cells. Acta Pharmacol Sin 28:459–472. https://doi.org/10.1111/j.1745-7254.2007.00549.x
Peng S, Yao J, Liu Y et al (2015) Activation of Nrf2 target enzymes conferring protection against oxidative stress in PC12 cells by ginger principal constituent 6-shogaol. Food Funct 6:2813–2823. https://doi.org/10.1039/C5FO00214A
Petri S, Korner S, Kiaei M (2012) Nrf2/ARE signaling pathway: key mediator in oxidative stress and potential therapeutic target in ALS. Neurol Res Int 2012:1–7. https://doi.org/10.1155/2012/878030
Poljsak B, Suput D, Milisav I (2013) Achieving the balance between ROS and antioxidants: when to use the synthetic antioxidants. Oxidative Med Cell Longev 2013:1–11. https://doi.org/10.1155/2013/956792
Prastowo S, Amin A, Rings F et al (2016) Fateful triad of reactive oxygen species, mitochondrial dysfunction and lipid accumulation is associated with expression outline of the AMP-activated protein kinase pathway in bovine blastocysts. Reprod Fertil Dev 49:193–202. https://doi.org/10.1071/RD15319
Qin W-S, Deng Y-H, Cui F-C (2016) Sulforaphane protects against acrolein-induced oxidative stress and inflammatory responses: modulation of Nrf-2 and COX-2 expression. Arch Med Sci 12:871–880. https://doi.org/10.5114/aoms.2016.59919
Saeed-Zidane M, Linden L, Salilew-Wondim D et al (2017) Cellular and exosome mediated molecular defense mechanism in bovine granulosa cells exposed to oxidative stress. PLoS One 12(11):e0187569. https://doi.org/10.1371/journal.pone.0187569
Shkolnik K, Tadmor A, Ben-Dor S et al (2011) Reactive oxygen species are indispensable in ovulation. Proc Natl Acad Sci U S A 108:1462–1467. https://doi.org/10.1073/pnas.1017213108
Sohel MMH, Cinar MU (2015) Advancement in molecular genetics to understand the molecular reproduction of livestock—follicular development. Res Agric Livest Fish 1:47–60. https://doi.org/10.3329/ralf.v1i1.22355
Sohel MMH, Hoelker M, Noferesti SS et al (2013) Exosomal and non-exosomal transport of extra-cellular microRNAs in follicular fluid: implications for bovine oocyte developmental competence. PLoS One. https://doi.org/10.1371/journal.pone.0078505
Sohel MH, Cinar MU, Kalibar M et al (2016) Appropriate concentration of hydrogen peroxide and sulforaphane for granulosa cells to study oxidative stress in vitro. J Biotechnol 231:S24. https://doi.org/10.1016/j.jbiotec.2016.05.104
Sohel MMH, Konca Y, Akyuz B et al (2017) Concentration dependent antioxidative and apoptotic effects of sulforaphane on bovine granulosa cells in vitro. Theriogenology 97:17–26. https://doi.org/10.1016/j.theriogenology.2017.04.015
Strober W (2001) Trypan blue exclusion test of cell viability. Curr Protoc Immunol Appendix 3:Appendix 3B. https://doi.org/10.1002/0471142735.ima03bs21
Visalli G, Facciolà A, Bertuccio MP et al (2017) In vitro assessment of the indirect antioxidant activity of Sulforaphane in redox imbalance vanadium-induced. Nat Prod Res 31:2612–2620. https://doi.org/10.1080/14786419.2017.1286485
Yada H, Hosokawa K, Tajima K et al (1999) Role of ovarian theca and granulosa cell interaction in hormone production and cell growth during the bovine follicular maturation process. Biol Reprod 61:1480–1486
Zhang M, An C, Gao Y et al (2013) Emerging roles of Nrf2 and phase II antioxidant enzymes in neuroprotection. Prog Neurobiol 100:30–47. https://doi.org/10.1016/j.pneurobio.2012.09.003
Acknowledgements
The authors would like to thank Mr. Heinz Biörnsen and Mr. Mahmut Kaliber for their assistance during sample collection.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Statement on the welfare of animals
This article does not contain any studies with live animals performed by any of the authors.
Additional information
The original version of this article was revised: There is an error in the Original Publication of this paper. Figures 1-6 were unupdated, thus corrected figures provided below:
Electronic supplementary material
Fig. S1
The purity of GCs. GCs were collected according to the protocol mentioned in the “Materials and methods” section. Specific primers targeting FSHR, CYP17A1 and GAPDH were designed using primer3web version 4.0.4 (http://bioinfo.ut.ee/primer3/). Primer sequences are available in Supplementary Table 1. The polymerase chain reaction (PCR) was set in a thermocycler with thermal cycle conditions of pre-incubation at 95°C for 5 min, 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C FSHR, 56°C GAPDH and 57°C CYP17A1 for 30 s, extension at 72°C for 1 min and a final extension at 72°C for 10 min. The corresponding PCR products were mixed with loading buffer and loaded into 2% agarose gel stained with Ethidium bromide (EtBr) and visualized using Chemidoc XRS (Bio-Rad) instrument (BIO-RAD, München, Germany) to detect the presence or absence of gene-specific bands. The results showed that the GC specific marker gene FSHR was detected at a higher level as indicated by a strong band, while no expression was detected for the CYP17A1 gene in GCs. These results confirmed that the isolation procedure of GCs was completely free of contamination from thecal cells. (PDF 117 kb)
Fig. S2
Expression of NRF2, KEAP1 and CASP3 in response to 10 μM SFN at different time points. Approximately 6 × 105 viable pure GCs were seeded in each well of a 6-well plate and grown until 40-50% confluence. Cells were then treated with 10 μM SFN for different time points (6 h, 12 h, 24 and 48 h) to identify the appropriate exposure time of SFN. We checked three different genes; NRF2 (inducer of the NRF2 antioxidant pathway), KEAP1 (negative controller of NRF2) and CASP3 (an indicator of apoptosis induction). The results showed that the expression of NRF2 was significantly higher in all exposure time points compared to the 6 h time point, however, the highest expression was observed in the 12 h and 24 h time points. On the other hand, the expression of KEAP1 was more or less similar across the time points. In addition, the expression of CASP3 was similar to control at 12 h and 24 h, however, it significantly increased at the 48 h time point. The expression analysis of these genes revealed that both the 12 h and 24 h exposure times are suitable for activating the antioxidant defense system without creating any damage to GCs. As most of the mammalian cell cycle is completed within 20-24 h, we decided to expose the GCs against 10 μM SFN for 24 h. (PDF 154 kb)
Fig. S3
Normality test of qPCR data. Normality of the qPCR data (Ct value) was tested using Microsoft Excel 2016. For this, all Ct values were organized in ascending order (smallest to largest). Afterwards, the cumulative distribution factor (CDF) was calculated using the 1 / (2*total count) formula for the first number, CDF1+2 / (2*total count) for the second number, CDF2+2 / (2*total count) for the third number and so on. Expected value was calculated using the NORM.INV(CDF, mean, standard deviation) formula and Z-score was calculated using the NORM.S.INV(CDF) formula. Normal probability plot for the Ct values shows that the data appear to be very close to being normally distributed. The actual Ct values (red) match very closely with the normally distributed (blue) Ct values that clearly indicate the data are normally distributed. (PDF 161 kb)
Table S1
(PDF 91.4 kb)
Rights and permissions
About this article
Cite this article
Sohel, M.M.H., Amin, A., Prastowo, S. et al. Sulforaphane protects granulosa cells against oxidative stress via activation of NRF2-ARE pathway. Cell Tissue Res 374, 629–641 (2018). https://doi.org/10.1007/s00441-018-2877-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-018-2877-z