Abstract
General transcription factor IIIC subunit 5 (GTF3C5) encodes transcription factor IIIC63 (TFIIIC63). It binds to DNA to recruit another transcription factor, TFIIIB, and RNA polymerase III (Pol III) to mediate the transcription of small noncoding RNAs, such as tRNAs. Here, we report four individuals from three families presenting with a multisystem developmental disorder phenotype with biallelic variants in GTF3C5. The overlapping features include growth retardation, developmental delay, intellectual disability, dental anomalies, cerebellar malformations, delayed bone age, skeletal anomalies, and facial dysmorphism. Using lymphoblastoid cell lines (LCLs) from two affected individuals, we observed a reduction in TFIIIC63 protein levels compared to control LCLs. Genome binding of TFIIIC63 protein is also reduced in LCL from one of the affected individuals. Additionally, approximately 40% of Pol III binding regions exhibited reduction in the level of Pol III occupancy in the mutant genome relative to the control, while approximately 54% of target regions showed comparable levels of Pol III occupancy between the two, indicating partial impairment of Pol III occupancy in the mutant genome. Yeasts with subject-specific variants showed temperature sensitivity and impaired growth, supporting the notion that the identified variants have deleterious effects. gtf3c5 mutant zebrafish showed developmental defects, including a smaller body, head, and eyes. Taken together, our data show that GTF3C5 plays an important role in embryonic development, and that biallelic variants in this gene cause a multisystem developmental disorder. Our study adds GTF3C5-related disorder to the growing list of genetic disorders associated with Pol III transcription machinery.






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Anazi S, Maddirevula S, Salpietro V et al (2017) Expanding the genetic heterogeneity of intellectual disability. Hum Genet 136:1419–1429. https://doi.org/10.1007/s00439-017-1843-2
Bernard G, Chouery E, Putorti ML et al (2011) Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. Am J Hum Genet 89:415–423. https://doi.org/10.1016/j.ajhg.2011.07.014
Borck G, Hög F, Dentici ML et al (2015) BRF1 mutations alter RNA polymerase III-dependent transcription and cause neurodevelopmental anomalies. Genome Res 25:155–166. https://doi.org/10.1101/gr.176925.114
Burke D, Dawson D, Stearns T, Cold Spring Harbor Laboratory (2000) Methods in yeast genetics : a cold spring harbor laboratory course manual, 2000 ed. Cold Spring Harbor Laboratory Press, Plainview
Dittmar KA, Goodenbour JM, Pan T (2006) Tissue-specific differences in human transfer RNA expression. PLoS Genet 2:e221. https://doi.org/10.1371/journal.pgen.0020221
Djordjevic D, Pinard M, Gauthier MS et al (2021) De novo variants in POLR3B cause ataxia, spasticity, and demyelinating neuropathy. Am J Hum Genet 108:186–193. https://doi.org/10.1016/J.AJHG.2020.12.002
Dumay-Odelot H, Durrieu-Gaillard S, Da Silva D et al (2010) Cell growth- and differentiation-dependent regulation of RNA polymerase III transcription. Cell Cycle 9:3711–3723. https://doi.org/10.4161/CC.9.18.13203
Goodfellow SJ, White RJ (2007) Regulation of RNA polymerase III transcription during mammalian cell growth. Cell Cycle 6:2323–2326. https://doi.org/10.4161/CC.6.19.4767
Honda S, Shigematsu M, Morichika K et al (2015) Four-leaf clover qRT-PCR: a convenient method for selective quantification of mature tRNA. RNA Biol 12:501–508. https://doi.org/10.1080/15476286.2015.1031951
Honjo RS, Castro MAA, Ferraciolli SF et al (2021) Cerebellofaciodental syndrome in an adult patient: expanding the phenotypic and natural history characteristics. Am J Med Genet A 185:1561–1568. https://doi.org/10.1002/AJMG.A.62140
Hsieh YJ, Wang Z, Kovelman R, Roeder RG (1999) Cloning and characterization of two evolutionarily conserved subunits (TFIIIC102 and TFIIIC63) of human TFIIIC and their involvement in functional interactions with TFIIIB and RNA polymerase III. Mol Cell Biol 19:4944–4952. https://doi.org/10.1128/mcb.19.7.4944
Iossifov I, O’Roak BJ, Sanders SJ et al (2014) The contribution of de novo coding mutations to autism spectrum disorder. Nature 515:216–221. https://doi.org/10.1038/NATURE13908
Jay AM, Conway RL, Thiffault I et al (2016) Neonatal progeriod syndrome associated with biallelic truncating variants in POLR3A. Am J Med Genet A 170:3343–3346. https://doi.org/10.1002/ajmg.a.37960
Jee YH, Sowada N, Markello TC et al (2017) BRF1 mutations in a family with growth failure, markedly delayed bone age, and central nervous system anomalies. Clin Genet 91:739–747. https://doi.org/10.1111/cge.12887
Jiang Y, Huang J, Tian K et al (2022) Cross-regulome profiling of RNA polymerases highlights the regulatory role of polymerase III on mRNA transcription by maintaining local chromatin architecture. Genome Biol 23:246. https://doi.org/10.1186/s13059-022-02812-w
Jourdain S, Acker J, Ducrot C et al (2003) The tau95 subunit of yeast TFIIIC influences upstream and downstream functions of TFIIIC.DNA complexes. J Biol Chem 278:10450–10457. https://doi.org/10.1074/JBC.M213310200
Karaca E, Harel T, Pehlivan D et al (2015) Genes that affect brain structure and function identified by rare variant analyses of mendelian neurologic disease. Neuron 88:499–513. https://doi.org/10.1016/j.neuron.2015.09.048
Karczewski KJ, Francioli LC, Tiao G et al (2020) The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581:434–443. https://doi.org/10.1038/s41586-020-2308-7
Khanna T, Hanna G, Sternberg MJE, David A (2021) Missense3D-DB web catalogue: an atom-based analysis and repository of 4M human protein-coding genetic variants. Hum Genet 140:805–812. https://doi.org/10.1007/S00439-020-02246-Z
Komata M, Katou Y, Tanaka H et al (2014) Chromatin immunoprecipitation protocol for mammalian cells. Methods Mol Biol 1164:33–38. https://doi.org/10.1007/978-1-4939-0805-9_4
Kosmicki JA, Samocha KE, Howrigan DP et al (2017) Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet 49:504–510. https://doi.org/10.1038/NG.3789
La PR, Tonduti D, Dressman HG et al (2014) Brain magnetic resonance imaging (MRI) pattern recognition in Pol III-related leukodystrophies. J Child Neurol 29:214–220. https://doi.org/10.1177/0883073813503902
Lata E, Choquet K, Sagliocco F et al (2021) RNA polymerase III subunit mutations in genetic diseases. Front Mol Biosci 8:752. https://doi.org/10.3389/FMOLB.2021.696438/BIBTEX
Longtine MS, McKenzie A, Demarini DJ et al (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961. https://doi.org/10.1002/(sici)1097-0061(199807)14:10%3c953::aid-yea293%3e3.0.co;2-u
Nakato R, Itoh T, Shirahige K (2013) DROMPA: easy-to-handle peak calling and visualization software for the computational analysis and validation of ChIP-seq data. Genes Cells 18:589–601. https://doi.org/10.1111/GTC.12058
Oler AJ, Alla RK, Roberts DN et al (2010) Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol 17:620–628. https://doi.org/10.1038/nsmb.1801
Paolacci S, Bertola D, Franco J et al (2017) Wiedemann-Rautenstrauch syndrome: a phenotype analysis. Am J Med Genet A 173:1763–1772. https://doi.org/10.1002/AJMG.A.38246
Paolacci S, Li Y, Agolini E et al (2018) Specific combinations of biallelic POLR3A variants cause Wiedemann–Rautenstrauch syndrome. J Med Genet 55:837–846. https://doi.org/10.1136/jmedgenet-2018-105528
Papuc SM, Abela L, Steindl K et al (2019) The role of recessive inheritance in early-onset epileptic encephalopathies: a combined whole-exome sequencing and copy number study. Eur J Hum Genet 27:408–421. https://doi.org/10.1038/S41431-018-0299-8
Rentas S, Rathi KS, Kaur M et al (2020) Diagnosing Cornelia de Lange syndrome and related neurodevelopmental disorders using RNA sequencing. Genet Med 22:927–936. https://doi.org/10.1038/S41436-019-0741-5
Reuter MS, Tawamie H, Buchert R et al (2017) Diagnostic yield and novel candidate genes by exome sequencing in 152 consanguineous families with neurodevelopmental disorders. JAMA Psychiat 74:293–299. https://doi.org/10.1001/jamapsychiatry.2016.3798
Saida K, Chong PF, Yamaguchi A et al (2022) Monogenic causes of pigmentary mosaicism. Hum Genet 141:1771–1784. https://doi.org/10.1007/S00439-022-02437-W
Saitsu H, Osaka H, Sasaki M et al (2011) Mutations in POLR3A and POLR3B encoding RNA polymerase III subunits cause an autosomal-recessive hypomyelinating leukoencephalopathy. Am J Hum Genet 89:644–651. https://doi.org/10.1016/j.ajhg.2011.10.003
Sharma P, Grabowski TB, Patiño R (2016) Thyroid endocrine disruption and external body morphology of Zebrafish. Gen Comp Endocrinol 226:42–49. https://doi.org/10.1016/j.ygcen.2015.12.023
Sobreira N, Schiettecatte F, Valle D, Hamosh A (2015) GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat 36:928–930. https://doi.org/10.1002/HUMU.22844
Taylor NMI, Baudin F, Von Scheven G, Müller CW (2013) RNA polymerase III-specific general transcription factor IIIC contains a heterodimer resembling TFIIF Rap30/Rap74. Nucl Acids Res 41:9183–9196. https://doi.org/10.1093/NAR/GKT664
Tétreault M, Choquet K, Orcesi S et al (2011) Recessive mutations in POLR3B, encoding the second largest subunit of Pol III, cause a rare hypomyelinating leukodystrophy. Am J Hum Genet 89:652–655. https://doi.org/10.1016/j.ajhg.2011.10.006
Wambach JA, Wegner DJ, Patni N et al (2018) Bi-allelic POLR3A loss-of-function variants cause autosomal-recessive wiedemann-rautenstrauch syndrome. Am J Hum Genet. https://doi.org/10.1016/j.ajhg.2018.10.010
Wang Z, Zhao G, Zhu Z et al (2024) VarCards2: an integrated genetic and clinical database for ACMG-AMP variant-interpretation guidelines in the human whole genome. Nucleic Acids Res. https://doi.org/10.1093/NAR/GKAD1061
Watt KE, Macintosh J, Bernard G, Trainor PA (2023) RNA Polymerases I and III in development and disease. Semin Cell Dev Biol 136:49–63. https://doi.org/10.1016/J.SEMCDB.2022.03.027
White RJ (2004) RNA polymerase III transcription and cancer. Oncogene 23:3208–3216. https://doi.org/10.1038/sj.onc.1207547
Winter AG, Sourvinos G, Allison SJ et al (2000) RNA polymerase III transcription factor TFIIIC2 is overexpressed in ovarian tumors. Proc Natl Acad Sci USA 97:12619–12624. https://doi.org/10.1073/PNAS.230224097/ASSET/104B505D-B427-4EEF-89C0-249A6B2E1593/ASSETS/GRAPHIC/PQ2302240006.JPEG
Wu L, Pan J, Thoroddsen V et al (2003) Novel small-molecule inhibitors of RNA polymerase III. Eukaryot Cell 2:256–264. https://doi.org/10.1128/EC.2.2.256-264.2003
Yoshimura A, Sutani T, Shirahige K (2021) Functional control of Eco1 through the MCM complex in sister chromatid cohesion. Gene 784:145584. https://doi.org/10.1016/J.GENE.2021.145584
Acknowledgements
We are exceptionally grateful to the individuals and families who participated in this research study. We appreciate the GeneMatcher website (Sobreira et al. 2015) for facilitating the collaboration. We thank Jill Murrell Ph.D of Division of Genomic Diagnostics at Children’s Hospital of Philadelphia for assistance with variant submission to ClinVar.
Funding
KI was supported by the Children’s Hospital of Philadelphia Research Institute Institutional Developmental Fund, the Children’s Hospital of Philadelphia Junior Faculty Pilot Grant, the Children’s Hospital of Philadelphia Foerderer award, the Children’s Hospital of Philadelphia Pathology Support, and the Children’s Hospital of Philadelphia Roberts Collaborative. KS and KI were supported by Grant-in-Aid for Transformative Research Areas (A) 20H05940 (Japan Society for the Promotion of Science). KS was supported by Grant-in-Aid for Scientific Research (S) 20H05686 (Japan Society for the Promotion of Science) and CREST JPMJCR18S5 (Japan Science and Technology Agency). AI-O was partly supported by T32GM008638 and the Children’s Hospital of Philadelphia Roberts Collaborative. YK was supported by Japan Society for the Promotion of Science Overseas Research Fellowship.
Author information
Authors and Affiliations
Contributions
Aiko Iwata-Otsubo and Kosuke Izumi conceptualized and designed the study. Experiments and data analyses were performed by Aiko Iwata-Otsubo, Atsunori Yoshimura, Toyonori Sakata, Sarah K. Fiordaliso, Christoph Seiler, Katsuhiko Shirahige, and Kosuke Izumi. Data collection was performed by Cara M. Skraban, Cesar Augusto P Alves, Yukiko Kuroda, Jaime Vengoechea, Angela Grochowsky, Paige Ernste, Lauren Lulis, Addie Nesbitt, Ahmad Abou Tayoun, Christopher Gray, Meghan C Towne, Kelly Radtke, Elizabeth A Normand, Lindsay Rhodes, and Kosuke Izumi. The first draft of the manuscript was written by Aiko Iwata-Otsubo and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
MCT and KR are employees of Ambry Genetics. EAN and LR are employees of GeneDx, LLC.
Conflict of interest
The following authors have no conflicts of interest to declare for this manuscript: AI-O,CMS, AY, TS, CAPA, SKF, YK, JV, AG, PE, LL, AN, AAT, CG, CS, KS, and KI. MCT and KR are employees of Ambry Genetics. EAN and LR are employees of GeneDx, LLC.
Consent to participate
Informed consent was obtained from Subjects 1 and 2. De-identified information was collected for Subjects 3 and 4.
Consent to publish
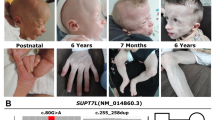
The authors affirm that human research participants provided informed consent for publication of the images in Figs. 1c, d.
Ethics approval
Subjects 1 and 2 were enrolled in the research study under an institutional review board-approved protocol (IRB16-013231). The animal study adhered to the principles set out in the Declaration of Helsinki. Animal experiments were approved by the Institutional Animal Care & Use Committees (IACUC) of the Children’s Hospital of Philadelphia (#21–001154).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Iwata-Otsubo, A., Skraban, C.M., Yoshimura, A. et al. Biallelic variants in GTF3C5, a regulator of RNA polymerase III-mediated transcription, cause a multisystem developmental disorder. Hum. Genet. 143, 437–453 (2024). https://doi.org/10.1007/s00439-024-02656-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-024-02656-3