Abstract
Mitochondrial disorders are collectively common, genetically heterogeneous disorders in both pediatric and adult populations. They are caused by molecular defects in oxidative phosphorylation, failure of essential bioenergetic supply to mitochondria, and apoptosis. Here, we present three affected individuals from a consanguineous family of Pakistani origin with variable seizures and intellectual disability. Both females display primary ovarian insufficiency (POI), while the male shows abnormal sex hormone levels. We performed whole exome sequencing and identified a recessive missense variant c.694C > T, p.Arg232Cys in TFAM that segregates with disease. TFAM (mitochondrial transcription factor A) is a component of the mitochondrial replisome machinery that maintains mtDNA transcription and replication. In primary dermal fibroblasts, we show depletion of mtDNA and significantly altered mitochondrial function and morphology. Moreover, we observed reduced nucleoid numbers with significant changes in nucleoid size or shape in fibroblasts from an affected individual compared to controls. We also investigated the effect of tfam impairment in zebrafish; homozygous tfam mutants carrying an in-frame c.141_149 deletion recapitulate the mtDNA depletion and ovarian dysgenesis phenotypes observed in affected humans. Together, our genetic and functional data confirm that TFAM plays a pivotal role in gonad development and expands the repertoire of mitochondrial disease phenotypes.






Similar content being viewed by others
Availability of data and material
Data, reagents, and mutant animal lines are available upon request and contingent upon appropriate ethics protocols from the requesting investigator and institution.
Code availability
Not applicable. Publicly available software was used for data analysis.
References
Alston CL, Rocha MC, Lax NZ, Turnbull DM, Taylor RW (2017) The genetics and pathology of mitochondrial disease. J Pathol 241:236–250. https://doi.org/10.1002/path.4809
Barshad G, Marom S, Cohen T, Mishmar D (2018) Mitochondrial DNA Transcription and Its Regulation: An Evolutionary Perspective. Trends Genet 34:682–692. https://doi.org/10.1016/j.tig.2018.05.009
Bavister BD, Squirrell JM (2000) Mitochondrial distribution and function in oocytes and early embryos. Hum Reprod 15(Suppl 2):189–198. https://doi.org/10.1093/humrep/15.suppl_2.189
Bonekamp NA, Larsson NG (2018) SnapShot: Mitochondrial Nucleoid. Cell 172(388–388):e1. https://doi.org/10.1016/j.cell.2017.12.039
Bourgeron T, Rustin P, Chretien D, Birch-Machin M, Bourgeois M, Viegas-Pequignot E, Munnich A, Rotig A (1995) Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat Genet 11:144–149. https://doi.org/10.1038/ng1095-144
Burbulla LF, Kruger R (2012) The use of primary human fibroblasts for monitoring mitochondrial phenotypes in the field of Parkinson’s disease. J vis Exp. https://doi.org/10.3791/4228
Campbell CT, Kolesar JE, Kaufman BA (2012) Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta 1819:921–929. https://doi.org/10.1016/j.bbagrm.2012.03.002
Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, Li Y, Ye J, Yu C, Li Z, Zhang X, Wang J, Yang H, Fang L, Chen Q (2018) SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 7:1–6. https://doi.org/10.1093/gigascience/gix120
Cox L, Liu JH (2014) Primary ovarian insufficiency: an update. Int J Womens Health 6:235–243. https://doi.org/10.2147/IJWH.S37636
Crowder CM, Lassiter CS, Gorelick DA (2018) Nuclear androgen receptor regulates testes organization and oocyte maturation in zebrafish. Endocrinology 159:980–993. https://doi.org/10.1210/en.2017-00617
Dairaghi DJ, Shadel GS, Clayton DA (1995) Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J Mol Biol 249:11–28. https://doi.org/10.1006/jmbi.1995.9889
Del Dotto V, Ullah F, Di Meo I, Magini P, Gusic M, Maresca A, Caporali L, Palombo F, Tagliavini F, Baugh EH, Macao B, Szilagyi Z, Peron C, Gustafson MA, Khan K, La Morgia C, Barboni P, Carbonelli M, Valentino ML, Liguori R, Shashi V, Sullivan J, Nagaraj S, El-Dairi M, Iannaccone A, Cutcutache I, Bertini E, Carrozzo R, Emma F, Diomedi-Camassei F, Zanna C, Armstrong M, Page M, Stong N, Boesch S, Kopajtich R, Wortmann S, Sperl W, Davis EE, Copeland WC, Seri M, Falkenberg M, Prokisch H, Katsanis N, Tiranti V, Pippucci T, Carelli V (2020) SSBP1 mutations cause mtDNA depletion underlying a complex optic atrophy disorder. J Clin Invest 130:108–125. https://doi.org/10.1172/JCI128514
Dimitromanolakis A, Paterson AD, Sun L (2019) Fast and accurate shared segment detection and relatedness estimation in un-phased genetic data via TRUFFLE. Am J Hum Genet 105:78–88. https://doi.org/10.1016/j.ajhg.2019.05.007
El-Hattab AW, Craigen WJ, Scaglia F (2017) Mitochondrial DNA maintenance defects. Biochim Biophys Acta Mol Basis Dis 1863:1539–1555. https://doi.org/10.1016/j.bbadis.2017.02.017
Ellis JL, Yin C (2017) Histological analyses of acute alcoholic liver injury in zebrafish. J vis Exp. https://doi.org/10.3791/55630
Fisher RP, Clayton DA (1985) A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavy- and light-strand promoters dissected and reconstituted in vitro. J Biol Chem 260:11330–11338
Halestrap AP, Price NT (1999) The proton-linked monocarboxylate transporter (MCT) family: structure, function and regulation. Biochem J 343(Pt 2):281–299
Hu Z, Ai N, Chen W, Wong QW, Ge W (2019) Loss of growth hormone gene (gh1) in zebrafish arrests folliculogenesis in females and delays spermatogenesis in males. Endocrinology 160:568–586. https://doi.org/10.1210/en.2018-00878
Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jakobs S (2011) Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci USA 108:13534–13539. https://doi.org/10.1073/pnas.1109263108
La Morgia C, Maresca A, Caporali L, Valentino ML, Carelli V (2020) Mitochondrial diseases in adults. J Intern Med 287:592–608. https://doi.org/10.1111/joim.13064
Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA (1998) Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet 18:231–236. https://doi.org/10.1038/ng0398-231
Laven JS (2016) Primary ovarian insufficiency. Semin Reprod Med 34:230–234. https://doi.org/10.1055/s-0036-1585402
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Longley MJ, Graziewicz MA, Bienstock RJ, Copeland WC (2005) Consequences of mutations in human DNA polymerase gamma. Gene 354:125–131. https://doi.org/10.1016/j.gene.2005.03.029
May-Panloup P, Chretien MF, Jacques C, Vasseur C, Malthiery Y, Reynier P (2005) Low oocyte mitochondrial DNA content in ovarian insufficiency. Hum Reprod 20:593–597. https://doi.org/10.1093/humrep/deh667
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110
Morino H, Pierce SB, Matsuda Y, Walsh T, Ohsawa R, Newby M, Hiraki-Kamon K, Kuramochi M, Lee MK, Klevit RE, Martin A, Maruyama H, King MC, Kawakami H (2014) Mutations in Twinkle primase-helicase cause Perrault syndrome with neurologic features. Neurology 83:2054–2061. https://doi.org/10.1212/WNL.0000000000001036
Ngo HB, Kaiser JT, Chan DC (2011) The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat Struct Mol Biol 18:1290–1296. https://doi.org/10.1038/nsmb.2159
Ngo HB, Lovely GA, Phillips R, Chan DC (2014) Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat Commun 5:3077. https://doi.org/10.1038/ncomms4077
Niederriter AR, Davis EE, Golzio C, Oh EC, Tsai IC, Katsanis N (2013) In vivo modeling of the morbid human genome using Danio rerio. J vis Exp. https://doi.org/10.3791/50338
Nunnari J, Suomalainen A (2012) Mitochondria: in sickness and in health. Cell 148:1145–1159. https://doi.org/10.1016/j.cell.2012.02.035
Otten ABC, Kamps R, Lindsey P, Gerards M, Pendeville-Samain H, Muller M, van Tienen FHJ, Smeets HJM (2020) Tfam knockdown results in reduction of mtDNA copy number, OXPHOS deficiency and abnormalities in zebrafish embryos. Front Cell Dev Biol 8:381. https://doi.org/10.3389/fcell.2020.00381
Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134:112–123. https://doi.org/10.1016/j.cell.2008.06.016
Perez GI, Trbovich AM, Gosden RG, Tilly JL (2000) Mitochondria and the death of oocytes. Nature 403:500–501. https://doi.org/10.1038/35000651
Quiros PM, Goyal A, Jha P, Auwerx J (2017) Analysis of mtDNA/nDNA ratio in mice. Curr Protoc Mouse Biol 7:47–54. https://doi.org/10.1002/cpmo.21
Rubio-Cosials A, Sidow JF, Jimenez-Menendez N, Fernandez-Millan P, Montoya J, Jacobs HT, Coll M, Bernado P, Sola M (2011) Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat Struct Mol Biol 18:1281–1289. https://doi.org/10.1038/nsmb.2160
Silva CA, Yamakami LY, Aikawa NE, Araujo DB, Carvalho JF, Bonfa E (2014) Autoimmune primary ovarian insufficiency. Autoimmun Rev 13:427–430. https://doi.org/10.1016/j.autrev.2014.01.003
Sobreira N, Schiettecatte F, Valle D, Hamosh A (2015) GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat 36:928–930. https://doi.org/10.1002/humu.22844
Stenton SL, Prokisch H (2018) Advancing genomic approaches to the molecular diagnosis of mitochondrial disease. Essays Biochem 62:399–408. https://doi.org/10.1042/EBC20170110
Stenton SL, Prokisch H (2020) Genetics of mitochondrial diseases: Identifying mutations to help diagnosis. EBioMedicine 56:102784. https://doi.org/10.1016/j.ebiom.2020.102784
Stenton SL, Kremer LS, Kopajtich R, Ludwig C, Prokisch H (2020) The diagnosis of inborn errors of metabolism by an integrative “multi-omics” approach: a perspective encompassing genomics, transcriptomics, and proteomics. J Inherit Metab Dis 43:25–35. https://doi.org/10.1002/jimd.12130
Stiles AR, Simon MT, Stover A, Eftekharian S, Khanlou N, Wang HL, Magaki S, Lee H, Partynski K, Dorrani N, Chang R, Martinez-Agosto JA, Abdenur JE (2016) Mutations in TFAM, encoding mitochondrial transcription factor A, cause neonatal liver failure associated with mtDNA depletion. Mol Genet Metab 119:91–99. https://doi.org/10.1016/j.ymgme.2016.07.001
Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wagele B, Altmaier E, CardioGram DP, Erdmann J, Grundberg E, Hammond CJ, de Angelis MH, Kastenmuller G, Kottgen A, Kronenberg F, Mangino M, Meisinger C, Meitinger T, Mewes HW, Milburn MV, Prehn C, Raffler J, Ried JS, Romisch-Margl W, Samani NJ, Small KS, Wichmann HE, Zhai G, Illig T, Spector TD, Adamski J, Soranzo N, Gieger C (2011) Human metabolic individuality in biomedical and pharmaceutical research. Nature 477:54–60. https://doi.org/10.1038/nature10354
Tan J, Wagner M, Stenton SL, Strom TM, Wortmann SB, Prokisch H, Meitinger T, Oexle K, Klopstock T (2020) Lifetime risk of autosomal recessive mitochondrial disorders calculated from genetic databases. EBioMedicine 54:102730. https://doi.org/10.1016/j.ebiom.2020.102730
Tiosano D, Mears JA, Buchner DA (2019) Mitochondrial dysfunction in primary ovarian insufficiency. Endocrinology 160:2353–2366. https://doi.org/10.1210/en.2019-00441
Tucker E, Jaillard S, Sinclair A (2019) Genetics and genomics of primary ovarian insufficiency. In: Leung P, Qiao J (eds) Human reproductive and prenatal genetics. Academic Press Ltd-Elsevier Science Ltd, pp 427–445. https://doi.org/10.1016/B978-0-12-813570-9.00019-X
Tucker EJ, Rius R, Jaillard S, Bell K, Lamont PJ, Travessa A, Dupont J, Sampaio L, Dulon J, Vuillaumier-Barrot S, Whalen S, Isapof A, Stojkovic T, Quijano-Roy S, Robevska G, van den Bergen J, Hanna C, Simpson A, Ayers K, Thorburn DR, Christodoulou J, Touraine P, Sinclair AH (2020) Genomic sequencing highlights the diverse molecular causes of Perrault syndrome: a peroxisomal disorder (PEX6), metabolic disorders (CLPP, GGPS1), and mtDNA maintenance/translation disorders (LARS2, TFAM). Hum Genet 139:1325–1343. https://doi.org/10.1007/s00439-020-02176-w
Vafai SB, Mootha VK (2012) Mitochondrial disorders as windows into an ancient organelle. Nature 491:374–383. https://doi.org/10.1038/nature11707
Van Blerkom J, Davis PW, Lee J (1995) ATP content of human oocytes and developmental potential and outcome after in-vitro fertilization and embryo transfer. Hum Reprod 10:415–424. https://doi.org/10.1093/oxfordjournals.humrep.a135954
Vozarikova V, Kunova N, Bauer JA, Frankovsky J, Kotrasova V, Prochazkova K, Dzugasova V, Kutejova E, Pevala V, Nosek J, Tomaska L (2020) Mitochondrial HMG-box containing proteins: from biochemical properties to the roles in human diseases. Biomolecules. https://doi.org/10.3390/biom10081193
Wang D, Li GD, Fan Y, Zhang DF, Bi R, Yu XF, Long H, Li YY, Yao YG (2017) The mtDNA replication-related genes TFAM and POLG are associated with leprosy in Han Chinese from Southwest China. J Dermatol Sci 88:349–356. https://doi.org/10.1016/j.jdermsci.2017.09.001
Acknowledgements
We are grateful to all the members of the families reported in this study for their participation and support of our research. We thank Azita Sadeghpour for facilitating primary cell line establishment. We thank the Beijing Genomics Institute (BGI) staff for technical support in generating WES data. We thank Niki Mourtzi for generating jointly called vcf files and Marie Mooney for critical review of the manuscript.
Funding
This study was funded by the Higher Education Commission of Pakistan under the International research support initiative program (IRSIP program; to FU); an Australian National Health and Medical Research Council (NHMRC) program grant (1074258, AHS); an Australian Mito Foundation grant (EJT); NHMRC fellowships (1054432 EJT, 1062854 AHS); and the United States National Institutes of Health grant P50-HD028138 (EED). EED is the Ann Marie and Francis Klocke MD Research Scholar.
Author information
Authors and Affiliations
Contributions
FU, WR, NK, SB and EED contributed to the conception and design of the study. SK and MT performed clinical investigation and ascertainment of the biological specimens for family 1. SH conducted WES and bioinformatic filtering. FU and KK validated WES data from family 1. FU and KK performed in vitro studies of primary cells. FU generated and phenotyped zebrafish models. KB, VCO, TP, AS and ET contributed data from family 2. KMB performed IBD analysis. FU, KK and EED drafted the manuscript. All authors read the manuscript and approved the submitted version.
Corresponding author
Ethics declarations
Conflicts of interest
NK is a significant shareholder in Rescindo Therapeutics.
Ethics approval
Human subjects research was approved by the National Institute for Biotechnology and Genetic Engineering (NIBGE), Faisalabad, Pakistan, and Lurie Children’s’ Hospital of Chicago, IL, USA. All zebrafish experiments were approved by institutional animal care and use committees (IACUC) of Duke University and Northwestern University.
Consent to participate
Written informed consent was obtained from all willing research participants.
Consent to publication
Written informed consent was obtained from all willing research participants for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
439_2021_2380_MOESM2_ESM.pdf
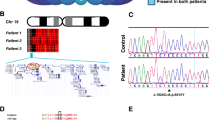
Figure S1. Homozygosity mapping shows a shared region between affected individuals in family 1. (A) Homozygous regions were mapped in affected individuals using HomozygosityMapper software. The homozygous blocks (red) were identified on chromosomes 6, 8 and 10 with >80% homozygosity. (B) Schematic representation of chromosome 10. TFAM is located on 10q21.2 (blue box). (C) Enlarged view of the chromosome 10 region shared between affected individuals (1-V-I and 1-V-II). (D) High resolution graphical representation of the homozygous region on chromosome 10 generated with GeneDistiller
439_2021_2380_MOESM3_ESM.pdf
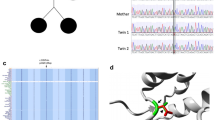
Figure S2. In silico protein modeling of TFAM p.Arg232Cys. Human TFAM protein sequence harboring either Arg232 (WT; top) or Cys232 (variant; bottom) was modeled in PyMOL and depicts TFAM (green) co-crystalized in a complex with light strand promoter (LSP; grey) sequence. The zoomed image (top right) shows Arg232 (cyan) interacting with Glu219 (red). Arginine changed to cysteine at position 232 may impair the interaction with glutamic acid at position 219 (bottom right) by disrupting the salt bridge present between these two amino acids in WT protein
439_2021_2380_MOESM4_ESM.pdf
Figure S3. CRISPR-Cas9 genome editing was used to target tfam in zebrafish. (A) Schematic shows the canonical transcript of tfam (GRCz11, ENSDART00000092009.6), green boxes, coding exons; untranslated regions (UTR), white boxes; black lines, introns. Exon targeted by sgRNA is shown with a black arrow. (B) Polyacrylamide gel image showing heteroduplex analysis of PCR products flanking the sgRNA target derived from two control embryos or embryos injected with tfam sgRNA plus Cas9 protein. Sizes (left) of the ladder are indicated in base pairs (bp). (C) Representative sequences generated from PCR products amplified from individual control or tfam F0 mutant larvae that were TOPO cloned. PAM, protospacer adjacent motif (blue box); sgRNA has an estimated 100% mosaicism (based on 12 colonies from 4 different embryos). (D) Representative sequence chromatogram confirms a 9 bp deletion in tfam homozygous mutants: c.141_149del p.(Lys48_Pro50del) that was isolated by outcrossing F0 adults to WT (ZDR), isolation of F1 mutants, and incrossing of heterozygous adults
439_2021_2380_MOESM5_ESM.pdf
Figure S4. tfam homozygous mutant zebrafish adults show morphological differences compared to WT. (A) Representative images of adult zebrafish siblings at 4 months age. WT type males and females can be readily distinguished but tfam homozygous mutants cannot be characterized visually as male or female. (B) Quantification of the body length of WT and tfam homozygous mutant siblings measured at 4 months of age. n=7-9/condition; ****p<0.0001; unpaired t-test; error bars indicate standard deviation
439_2021_2380_MOESM6_ESM.pdf
Figure S5. tfam homozygous mutant zebrafish display a depletion in mitochondrial complex transcripts and mtDNA content. (A, B) Quantification of mtDNA copy number shows a significant depletion in tfam homozygous mutant adult head and tail tissues, two biological replicates; A, n=2 animals; B, n=3 animals. (C) Quantification of mt-nd1 and mt-co1 by qRT-PCR shows a significant reduction in tfam homozygous mutant adult gonad tissue. Repeated once with similar results, technical triplicates. (A-B-C). ****p < 0.0001; ***p<0.001, unpaired t-test; error bars indicate standard deviation
439_2021_2380_MOESM7_ESM.pdf
Figure S6. Validation of the morpholino used to ablate tfam in zebrafish. (A) Schematic shows the canonical zebrafish tfam transcript (GRCz11, ENSDART00000092009.6), green boxes, coding exons; untranslated regions (UTR), white boxes; black lines, introns. Arrow indicates morpholino (MO) target site at the exon 3 splice donor. (B) Agarose gel images show an aberrantly spliced transcript induced by tfam MO; cDNA was generated from total RNA isolated from 2 dpf embryos. The expected wild-type fragment is 378 bp. b-actin to was used to control for RNA integrity. (C) Representative sequences generated from individual colonies that were TOPO cloned from RT-PCR products generated from control and tfam morphants; exon 3 is excluded, resulting in mRNA lacking 71 bp in morphants. (D) Quantification of mtDNA copy number shows a significant depletion in tfam F0 mutants and morphants, two biological replicates; n=25 larvae per condition
439_2021_2380_MOESM8_ESM.pdf
Figure S7. In vivo complementation assay suggests that p.Arg232Cys impairs TFAM function. Zebrafish embryos were injected with tfam MO in the presence or absence of human TFAM mRNA; aged to 4 dpf; and pools of 25 larvae were harvested for quantification of mtDNA copy number using mt-nd1 as a mitochondrial marker and globin as a reference for nuclear genomic DNA. p.Pro178Leu is known to be pathogenic and has an absolute mtDNA content indistinguishable from MO alone. *p<0.05; **p<0.01; ns, not significant; data were generated from two biological replicates and compared with an unpaired t-test; error bars indicate standard deviation
Rights and permissions
About this article
Cite this article
Ullah, F., Rauf, W., Khan, K. et al. A recessive variant in TFAM causes mtDNA depletion associated with primary ovarian insufficiency, seizures, intellectual disability and hearing loss. Hum Genet 140, 1733–1751 (2021). https://doi.org/10.1007/s00439-021-02380-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-021-02380-2