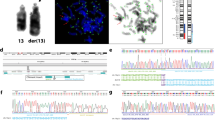
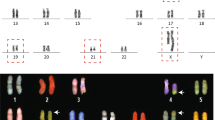
Abstract
Chromosomal insertions are thought to be rare structural rearrangements. The current understanding of the underlying mechanisms of their origin is still limited. In this study, we sequenced 16 cases with apparent simple insertions previously identified by karyotyping and/or chromosomal microarray analysis. Using mate-pair genome sequencing (GS), we identified all 16 insertions and revised previously designated karyotypes in 75.0% (12/16) of the cases. Additional cryptic rearrangements were identified in 68.8% of the cases (11/16). The incidence of additional cryptic rearrangements in chromosomal insertions was significantly higher compared to balanced translocations and inversions reported in other studies by GS. We characterized and classified the cryptic insertion rearrangements into four groups, which were not mutually exclusive: (1) insertion segments were fragmented and their subsegments rearranged and clustered at the insertion site (10/16, 62.5%); (2) one or more cryptic subsegments were not inserted into the insertion site (5/16, 31.3%); (3) segments of the acceptor chromosome were scattered and rejoined with the insertion segments (2/16, 12.5%); and (4) copy number gains were identified in the flanking regions of the insertion site (2/16, 12.5%). In addition to the observation of these chromothripsis- or chromoanasynthesis-like events, breakpoint sequence analysis revealed microhomology to be the predominant feature. However, no significant correlation was found between the number of cryptic rearrangements and the size of the insertion. Overall, our study provide molecular characterization of karyotypically apparent simple insertions, demonstrate previously underappreciated complexities, and evidence that chromosomal insertions are likely formed by nonhomologous end joining and/or microhomology-mediated replication-based DNA repair.





Similar content being viewed by others
Data availability
The genome sequencing data of five samples used in this study have been made available on the CNGB Nucleotide Sequence Archive (CNSA: https://db.cngb.org/cnsa) under the accession number CNP0000078. For the other 12 cases, raw sequencing data have not been made available because most subjects did not consent to such sharing.
Code availability
NA.
References
Abel HJ, Larson DE, Regier AA, Chiang C, Das I, Kanchi KL, Layer RM, Neale BM, Salerno WJ, Reeves C, Buyske S, Genomics NCfCD, Matise TC, Muzny DM, Zody MC, Lander ES, Dutcher SK, Stitziel NO, Hall IM (2020) Mapping and characterization of structural variation in 17,795 human genomes. Nature 583:83–89. https://doi.org/10.1038/s41586-020-2371-0
Ankala A, Kohn JN, Hegde A, Meka A, Ephrem CL, Askree SH, Bhide S, Hegde MR (2012) Aberrant firing of replication origins potentially explains intragenic nonrecurrent rearrangements within genes, including the human DMD gene. Genome Res 22:25–34. https://doi.org/10.1101/gr.123463.111
Bacolla A, Wang G, Jain A, Chuzhanova NA, Cer RZ, Collins JR, Cooper DN, Bohr VA, Vasquez KM (2011) Non-B DNA-forming sequences and WRN deficiency independently increase the frequency of base substitution in human cells. J Biol Chem 286:10017–10026. https://doi.org/10.1074/jbc.M110.176636
Bauters M, Van Esch H, Friez MJ, Boespflug-Tanguy O, Zenker M, Vianna-Morgante AM, Rosenberg C, Ignatius J, Raynaud M, Hollanders K, Govaerts K, Vandenreijt K, Niel F, Blanc P, Stevenson RE, Fryns JP, Marynen P, Schwartz CE, Froyen G (2008) Nonrecurrent MECP2 duplications mediated by genomic architecture-driven DNA breaks and break-induced replication repair. Genome Res 18:847–858. https://doi.org/10.1101/gr.075903.107
Cer RZ, Bruce KH, Donohue DE, Temiz NA, Mudunuri US, Yi M, Volfovsky N, Bacolla A, Luke BT, Collins JR, Stephens RM (2012) Searching for non-B DNA-forming motifs using nBMST (non-B DNA motif search tool). Curr Protoc Hum Genet 18(7):1–22. https://doi.org/10.1002/0471142905.hg1807s73
Chau MHK, Cao Y, Kwok YKY, Chan S, Chan YM, Wang H, Yang Z, Wong HK, Leung TY, Choy KW (2019) Characteristics and mode of inheritance of pathogenic copy number variants in prenatal diagnosis. Am J Obstet Gynecol 221:493.e1. https://doi.org/10.1016/j.ajog.2019.06.007
Chau MHK, Wang H, Lai Y, Zhang Y, Xu F, Tang Y, Wang Y, Chen Z, Leung TY, Chung JPW, Kwok YK, Chong SC, Choy KW, Zhu Y, Xiong L, Wei W, Dong Z (2020) Low-pass genome sequencing: a validated method in clinical cytogenetics. Hum Genet. https://doi.org/10.1007/s00439-020-02185-9
Choy KW, Wang H, Shi M, Chen J, Yang Z, Zhang R, Yan H, Wang Y, Chen S, Chau MHK, Cao Y, Chan OYM, Kwok YK, Zhu Y, Chen M, Leung TY, Dong Z (2019) Prenatal diagnosis of fetuses with increased nuchal translucency by genome sequencing analysis. Front Genet 10:761. https://doi.org/10.3389/fgene.2019.00761
Collins RL, Brand H, Redin CE, Hanscom C, Antolik C, Stone MR, Glessner JT, Mason T, Pregno G, Dorrani N, Mandrile G, Giachino D, Perrin D, Walsh C, Cipicchio M, Costello M, Stortchevoi A, An JY, Currall BB, Seabra CM, Ragavendran A, Margolin L, Martinez-Agosto JA, Lucente D, Levy B, Sanders SJ, Wapner RJ, Quintero-Rivera F, Kloosterman W, Talkowski ME (2017) Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol 18:36. https://doi.org/10.1186/s13059-017-1158-6
Collins RL, Brand H, Karczewski KJ, Zhao X, Alfoldi J, Francioli LC, Khera AV, Lowther C, Gauthier LD, Wang H, Watts NA, Solomonson M, O'Donnell-Luria A, Baumann A, Munshi R, Walker M, Whelan CW, Huang Y, Brookings T, Sharpe T, Stone MR, Valkanas E, Fu J, Tiao G, Laricchia KM, Ruano-Rubio V, Stevens C, Gupta N, Cusick C, Margolin L, Genome Aggregation Database Production T, Genome Aggregation Database C, Taylor KD, Lin HJ, Rich SS, Post WS, Chen YI, Rotter JI, Nusbaum C, Philippakis A, Lander E, Gabriel S, Neale BM, Kathiresan S, Daly MJ, Banks E, MacArthur DG, Talkowski ME (2020) A structural variation reference for medical and population genetics. Nature 581:444–451. https://doi.org/10.1038/s41586-020-2287-8
Cretu Stancu M, van Roosmalen MJ, Renkens I, Nieboer MM, Middelkamp S, de Ligt J, Pregno G, Giachino D, Mandrile G, Espejo Valle-Inclan J, Korzelius J, de Bruijn E, Cuppen E, Talkowski ME, Marschall T, de Ridder J, Kloosterman WP (2017) Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat Commun 8:1326. https://doi.org/10.1038/s41467-017-01343-4
David D, Freixo JP, Fino J, Carvalho I, Marques M, Cardoso M, Pina-Aguilar RE, Morton CC (2020) Comprehensive clinically oriented workflow for nucleotide level resolution and interpretation in prenatal diagnosis of de novo apparently balanced chromosomal translocations in their genomic landscape. Hum Genet 139:531–543. https://doi.org/10.1007/s00439-020-02121-x
de Pagter MS, van Roosmalen MJ, Baas AF, Renkens I, Duran KJ, van Binsbergen E, Tavakoli-Yaraki M, Hochstenbach R, van der Veken LT, Cuppen E, Kloosterman WP (2015) Chromothripsis in healthy individuals affects multiple protein-coding genes and can result in severe congenital abnormalities in offspring. Am J Hum Genet 96:651–656. https://doi.org/10.1016/j.ajhg.2015.02.005
Dong Z, Jiang L, Yang C, Hu H, Wang X, Chen H, Choy KW, Hu H, Dong Y, Hu B, Xu J, Long Y, Cao S, Chen H, Wang WJ, Jiang H, Xu F, Yao H, Xu X, Liang Z (2014) A robust approach for blind detection of balanced chromosomal rearrangements with whole-genome low-coverage sequencing. Hum Mutat 35:625–636. https://doi.org/10.1002/humu.22541
Dong Z, Zhang J, Hu P, Chen H, Xu J, Tian Q, Meng L, Ye Y, Wang J, Zhang M, Li Y, Wang H, Yu S, Chen F, Xie J, Jiang H, Wang W, Choy KW, Xu Z (2016) Low-pass whole-genome sequencing in clinical cytogenetics: a validated approach. Genet Med 18:940–948. https://doi.org/10.1038/gim.2015.199
Dong Z, Wang H, Chen H, Jiang H, Yuan J, Yang Z, Wang WJ, Xu F, Guo X, Cao Y, Zhu Z, Geng C, Cheung WC, Kwok YK, Yang H, Leung TY, Morton CC, Cheung SW, Choy KW (2018) Identification of balanced chromosomal rearrangements previously unknown among participants in the 1000 genomes project: implications for interpretation of structural variation in genomes and the future of clinical cytogenetics. Genet Med 20:697–707. https://doi.org/10.1038/gim.2017.170
Dong Z, Yan J, Xu F, Yuan J, Jiang H, Wang H, Chen H, Zhang L, Ye L, Xu J, Shi Y, Yang Z, Cao Y, Chen L, Li Q, Zhao X, Li J, Chen A, Zhang W, Wong HG, Qin Y, Zhao H, Chen Y, Li P, Ma T, Wang WJ, Kwok YK, Jiang Y, Pursley AN, Chung JPW, Hong Y, Kristiansen K, Yang H, Pina-Aguilar RE, Leung TY, Cheung SW, Morton CC, Choy KW, Chen ZJ (2019a) Genome sequencing explores complexity of chromosomal abnormalities in recurrent miscarriage. Am J Hum Genet 105:1102–1111. https://doi.org/10.1016/j.ajhg.2019.10.003
Dong Z, Zhao X, Li Q, Yang Z, Xi Y, Alexeev A, Shen H, Wang O, Ruan J, Ren H, Wei H, Qi X, Li J, Zhu X, Zhang Y, Dai P, Kong X, Kirkconnell K, Alferov O, Giles S, Yamtich J, Kermani BG, Dong C, Liu P, Mi Z, Zhang W, Xu X, Drmanac R, Choy KW, Jiang Y (2019b) Development of coupling controlled polymerizations by adapter-ligation in mate-pair sequencing for detection of various genomic variants in one single assay. DNA Res. https://doi.org/10.1093/dnares/dsz011
Ferraris A, Bernardini L, Avramovska VS, Zanni G, Loddo S, Sukarova-Angelovska E, Parisi V, Capalbo A, Tumini S, Travaglini L, Mancini F, Duma F, Barresi S, Novelli A, Mercuri E, Tarani L, Italian CSG, Bertini E, Dallapiccola B, Valente EM (2013) Dandy-Walker malformation and Wisconsin syndrome: novel cases add further insight into the genotype-phenotype correlations of 3q23q25 deletions. Orphanet J Rare Dis 8:75. https://doi.org/10.1186/1750-1172-8-75
Gu S, Yuan B, Campbell IM, Beck CR, Carvalho CM, Nagamani SC, Erez A, Patel A, Bacino CA, Shaw CA, Stankiewicz P, Cheung SW, Bi W, Lupski JR (2015) Alu-mediated diverse and complex pathogenic copy-number variants within human chromosome 17 at p13.3. Hum Mol Genet 24:4061–4077. https://doi.org/10.1093/hmg/ddv146
Gu S, Szafranski P, Akdemir ZC, Yuan B, Cooper ML, Magrina MA, Bacino CA, Lalani SR, Breman AM, Smith JL, Patel A, Song RH, Bi W, Cheung SW, Carvalho CM, Stankiewicz P, Lupski JR (2016) Mechanisms for complex chromosomal insertions. PLoS Genet 12:e1006446. https://doi.org/10.1371/journal.pgen.1006446
Hagens O, Dubos A, Abidi F, Barbi G, Van Zutven L, Hoeltzenbein M, Tommerup N, Moraine C, Fryns JP, Chelly J, van Bokhoven H, Gecz J, Dollfus H, Ropers HH, Schwartz CE, de Santos RCSD, Kalscheuer V, Hanauer A (2006) Disruptions of the novel KIAA1202 gene are associated with X-linked mental retardation. Hum Genet 118:578–590. https://doi.org/10.1007/s00439-005-0072-2
Hijazi H, Coelho FS, Gonzaga-Jauregui C, Bernardini L, Mar SS, Manning MA, Hanson-Kahn A, Naidu S, Srivastava S, Lee JA, Jones JR, Friez MJ, Alberico T, Torres B, Fang P, Cheung SW, Song X, Davis-Williams A, Jornlin C, Wight PA, Patyal P, Taube J, Poretti A, Inoue K, Zhang F, Pehlivan D, Carvalho CMB, Hobson GM, Lupski JR (2020) Xq22 deletions and correlation with distinct neurological disease traits in females: further evidence for a contiguous gene syndrome. Hum Mutat 41:150–168. https://doi.org/10.1002/humu.23902
Huang J, Poon LC, Akolekar R, Choy KW, Leung TY, Nicolaides KH (2014) Is high fetal nuchal translucency associated with submicroscopic chromosomal abnormalities on array CGH? Ultrasound Obstet Gynecol 43:620–624. https://doi.org/10.1002/uog.13384
Kang SH, Shaw C, Ou Z, Eng PA, Cooper ML, Pursley AN, Sahoo T, Bacino CA, Chinault AC, Stankiewicz P, Patel A, Lupski JR, Cheung SW (2010) Insertional translocation detected using FISH confirmation of array-comparative genomic hybridization (aCGH) results. Am J Med Genet A 152A:1111–1126. https://doi.org/10.1002/ajmg.a.33278
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O'Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, Genome Aggregation Database C, Neale BM, Daly MJ, MacArthur DG (2020) The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581:434–443. https://doi.org/10.1038/s41586-020-2308-7
Kato T, Ouchi Y, Inagaki H, Makita Y, Mizuno S, Kajita M, Ikeda T, Takeuchi K, Kurahashi H (2017) Genomic characterization of chromosomal insertions: insights into the mechanisms underlying chromothripsis. Cytogenet Genome Res 153:1–9. https://doi.org/10.1159/000481586
Kehrer M, Liehr T, Benkert T, Singer S, Grasshoff U, Schaeferhoff K, Bonin M, Weichselbaum A, Tzschach A (2015) Interstitial duplication of chromosome region 1q25.1q25.3: report of a patient with mild cognitive deficits, tall stature and facial dysmorphisms. Am J Med Genet A 167A:653–656. https://doi.org/10.1002/ajmg.a.36943
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, Bacino CA, Campos-Acevedo LD, Delgado MR, Freedenberg D, Garnica A, Grebe TA, Hernandez-Almaguer D, Immken L, Lalani SR, McLean SD, Northrup H, Scaglia F, Strathearn L, Trapane P, Kang SH, Patel A, Cheung SW, Hastings PJ, Stankiewicz P, Lupski JR, Bi W (2011) Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146:889–903. https://doi.org/10.1016/j.cell.2011.07.042
Logsdon GA, Vollger MR, Eichler EE (2020) Long-read human genome sequencing and its applications. Nat Rev Genet. https://doi.org/10.1038/s41576-020-0236-x
Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, Santos-Simarro F, Gilbert-Dussardier B, Wittler L, Borschiwer M, Haas SA, Osterwalder M, Franke M, Timmermann B, Hecht J, Spielmann M, Visel A, Mundlos S (2015) Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161:1012–1025. https://doi.org/10.1016/j.cell.2015.04.004
Mackie Ogilvie C, Scriven PN (2002) Meiotic outcomes in reciprocal translocation carriers ascertained in 3-day human embryos. Eur J Hum Genet 10:801–806. https://doi.org/10.1038/sj.ejhg.5200895
Melotte C, Debrock S, D'Hooghe T, Fryns JP, Vermeesch JR (2004) Preimplantation genetic diagnosis for an insertional translocation carrier. Hum Reprod 19:2777–2783. https://doi.org/10.1093/humrep/deh539
Mendez-Rosado LA, Lantigua A, Galarza J, Hamid Al-Rikabi AB, Ziegler M, Liehr T (2017) Unusual de novo partial trisomy 17p12p11.2 due to unbalanced insertion into 5p13.1 in a severely affected boy. J Pediatr Genet 6:165–168. https://doi.org/10.1055/s-0037-1599195
Neill NJ, Ballif BC, Lamb AN, Parikh S, Ravnan JB, Schultz RA, Torchia BS, Rosenfeld JA, Shaffer LG (2011) Recurrence, submicroscopic complexity, and potential clinical relevance of copy gains detected by array CGH that are shown to be unbalanced insertions by FISH. Genome Res 21:535–544. https://doi.org/10.1101/gr.114579.110
Ottaviani D, LeCain M, Sheer D (2014) The role of microhomology in genomic structural variation. Trends Genet 30:85–94. https://doi.org/10.1016/j.tig.2014.01.001
Pellestor F, Anahory T, Lefort G, Puechberty J, Liehr T, Hedon B, Sarda P (2011) Complex chromosomal rearrangements: origin and meiotic behavior. Hum Reprod Update 17:476–494. https://doi.org/10.1093/humupd/dmr010
Redin C, Brand H, Collins RL, Kammin T, Mitchell E, Hodge JC, Hanscom C, Pillalamarri V, Seabra CM, Abbott MA, Abdul-Rahman OA, Aberg E, Adley R, Alcaraz-Estrada SL, Alkuraya FS, An Y, Anderson MA, Antolik C, Anyane-Yeboa K, Atkin JF, Bartell T, Bernstein JA, Beyer E, Blumenthal I, Bongers EM, Brilstra EH, Brown CW, Bruggenwirth HT, Callewaert B, Chiang C, Corning K, Cox H, Cuppen E, Currall BB, Cushing T, David D, Deardorff MA, Dheedene A, D'Hooghe M, de Vries BB, Earl DL, Ferguson HL, Fisher H, FitzPatrick DR, Gerrol P, Giachino D, Glessner JT, Gliem T, Grady M, Graham BH, Griffis C, Gripp KW, Gropman AL, Hanson-Kahn A, Harris DJ, Hayden MA, Hill R, Hochstenbach R, Hoffman JD, Hopkin RJ, Hubshman MW, Innes AM, Irons M, Irving M, Jacobsen JC, Janssens S, Jewett T, Johnson JP, Jongmans MC, Kahler SG, Koolen DA, Korzelius J, Kroisel PM, Lacassie Y, Lawless W, Lemyre E, Leppig K, Levin AV, Li H, Li H, Liao EC, Lim C, Lose EJ, Lucente D, Macera MJ, Manavalan P, Mandrile G, Marcelis CL, Margolin L, Mason T, Masser-Frye D, McClellan MW, Mendoza CJ, Menten B, Middelkamp S, Mikami LR, Moe E, Mohammed S, Mononen T, Mortenson ME et al (2017) The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat Genet 49:36–45. https://doi.org/10.1038/ng.3720
Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal S, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Quail MA, Burton J, Swerdlow H, Carter NP, Morsberger LA, Iacobuzio-Donahue C, Follows GA, Green AR, Flanagan AM, Stratton MR, Futreal PA, Campbell PJ (2011) Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144:27–40. https://doi.org/10.1016/j.cell.2010.11.055
Van Hemel JO, Eussen HJ (2000) Interchromosomal insertions. Identification of five cases and a review. Hum Genet 107:415–432. https://doi.org/10.1007/s004390000398
Vissers LE, Bhatt SS, Janssen IM, Xia Z, Lalani SR, Pfundt R, Derwinska K, de Vries BB, Gilissen C, Hoischen A, Nesteruk M, Wisniowiecka-Kowalnik B, Smyk M, Brunner HG, Cheung SW, van Kessel AG, Veltman JA, Stankiewicz P (2009) Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum Mol Genet 18:3579–3593. https://doi.org/10.1093/hmg/ddp306
Wang H, Xu X (2017) Microhomology-mediated end joining: new players join the team. Cell Biosci 7:6. https://doi.org/10.1186/s13578-017-0136-8
Wang L, Xi Y, Zhang W, Wang W, Shen H, Wang X, Zhao X, Alexeev A, Peters BA, Albert A, Xu X, Ren H, Wang O, Kirkconnell K, Perazich H, Clark S, Hurowitz E, Chen A, Xu X, Drmanac R, Jiang Y (2019) 3' Branch ligation: a novel method to ligate non-complementary DNA to recessed or internal 3'OH ends in DNA or RNA. DNA Res 26:45–53. https://doi.org/10.1093/dnares/dsy037
Wang H, Dong Z, Zhang R, Chau MHK, Yang Z, Tsang KYC, Wong HK, Gui B, Meng Z, Xiao K, Zhu X, Wang Y, Chen S, Leung TY, Cheung SW, Kwok YK, Morton CC, Zhu Y, Choy KW (2020) Low-pass genome sequencing versus chromosomal microarray analysis: implementation in prenatal diagnosis. Genet Med 22:500–510. https://doi.org/10.1038/s41436-019-0634-7
Funding
This project is supported by the National Natural Science Foundation of China (31801042, 81741004 and 81741009), the Health and Medical Research Fund (04152666, 07180576), and Direct Grant (2019.051).
Author information
Authors and Affiliations
Contributions
SWC, ZD, MHKC, TYL and KWC designed the study. SWC and MHKC collected the samples. MHKC, YZ performed mate-pair sequencing. ZD, MHKC, PD, XK, PS and YK performed the analysis and data interpretation. MHKC, PD and YZ conducted the validation. ZD, MHKC, PS, SWC and KWC wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest or competing interest.
Ethics approval
The study was approved by the institutional review boards of each collaborative site.
Informed consent
Written informed consent for sample storage and genetic analyses of peripheral bloods and invasive diagnostic samples was obtained from each participant.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dong, Z., Chau, M.H.K., Zhang, Y. et al. Deciphering the complexity of simple chromosomal insertions by genome sequencing. Hum Genet 140, 361–380 (2021). https://doi.org/10.1007/s00439-020-02210-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-020-02210-x