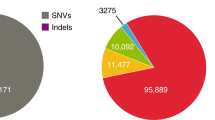
Abstract
An ethnicity is characterized by genomic fragments, single nucleotide polymorphisms (SNPs), and structural variations specific to it. However, the widely used ‘standard human reference genome’ GRCh37/38 is based on Caucasians. Therefore, de novo-assembled reference genomes for specific ethnicities would have advantages for genetics and precision medicine applications, especially with the long-read sequencing techniques that facilitate genome assembly. In this study, we assessed the de novo-assembled Chinese Han reference genome HX1 vis-à-vis the standard GRCh38 for improving the quality of assembly and for ethnicity-specific applications. Surprisingly, all genomic sequencing datasets mapped better to GRCh38 than to HX1, even for the datasets of the Chinese Han population. This gap was mainly due to the massive structural misassembly of the HX1 reference genome rather than the SNPs between the ethnicities, and this misassembly could not be corrected by short-read whole-genome sequencing (WGS). For example, HX1 and the other de novo-assembled personal genomes failed to assemble the mitochondrial genome as a contig. We mapped 97.1% of dbSNP, 98.8% of ClinVar, and 97.2% of COSMIC variants to HX1. HX1-absent, non-synonymous ClinVar SNPs were involved in 140 genes and many important functions in various diseases, most of which were due to the assembly failure of essential exons. In contrast, the HX1-specific regions were scantly expressible, as shown in the cell lines and clinical samples of Chinese patients. Our results demonstrated that the de novo-assembled individual genome such as HX1 did not have advantages against the standard GRCh38 genome due to insufficient assembly quality, and that it is, therefore, not recommended for common use.





Similar content being viewed by others
References
Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA, Genomes Project C (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. https://doi.org/10.1038/nature11632
Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR, Genomes Project C (2015) A global reference for human genetic variation. Nature 526:68–74
Bloom JS, Khan Z, Kruglyak L, Singh M, Caudy AA (2009) Measuring differential gene expression by short read sequencing: quantitative comparison to 2-channel gene expression microarrays. BMC Genomics 10:221. https://doi.org/10.1186/1471-2164-10-221
Cai N, Bigdeli TB, Kretzschmar WW, Li Y, Liang J, Hu J, Peterson RE, Bacanu S, Webb BT, Riley B, Li Q, Marchini J, Mott R, Kendler KS, Flint J (2017) 11,670 whole-genome sequences representative of the Han Chinese population from the CONVERGE project. Sci Data 4:170011. https://doi.org/10.1038/sdata.2017.11
Carlsson J, Gauthier DT, Carlsson JE, Coughlan JP, Dillane E, Fitzgerald RD, Keating U, McGinnity P, Mirimin L, Cross TF (2013) Rapid, economical single-nucleotide polymorphism and microsatellite discovery based on de novo assembly of a reduced representation genome in a non-model organism: a case study of Atlantic cod Gadus morhua. J Fish Biol 82:944–958. https://doi.org/10.1111/jfb.12034
Cho YS, Kim H, Kim HM, Jho S, Jun J, Lee YJ, Chae KS, Kim CG, Kim S, Eriksson A, Edwards JS, Lee S, Kim BC, Manica A, Oh TK, Church GM, Bhak J (2016) An ethnically relevant consensus Korean reference genome is a step towards personal reference genomes. Nat Commun 7:13637. https://doi.org/10.1038/ncomms13637
Dayama G, Emery SB, Kidd JM, Mills RE (2014) The genomic landscape of polymorphic human nuclear mitochondrial insertions. Nucleic Acids Res 42:12640–12649. https://doi.org/10.1093/nar/gku1038
Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S (2009) Real-time DNA sequencing from single polymerase molecules. Science 323:133–138. https://doi.org/10.1126/science.1162986
Gnerre S, Maccallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, Berlin AM, Aird D, Costello M, Daza R, Williams L, Nicol R, Gnirke A, Nusbaum C, Lander ES, Jaffe DB (2011) High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA 108:1513–1518. https://doi.org/10.1073/pnas.1017351108
Hindorff LA, Gillanders EM, Manolio TA (2011) Genetic architecture of cancer and other complex diseases: lessons learned and future directions. Carcinogenesis 32:945–954. https://doi.org/10.1093/carcin/bgr056
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921. https://doi.org/10.1038/35057062
Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Li S, Yang H, Wang J, Wang J (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272. https://doi.org/10.1101/gr.097261.109
Li D, Lu S, Liu W, Zhao X, Mai Z, Zhang G (2018) Optimal settings of mass spectrometry open search strategy for higher confidence. J Proteome Res 17:3719–3729. https://doi.org/10.1021/acs.jproteome.8b00352
Liu W, Xiang L, Zheng T, Jin J, Zhang G (2018) TranslatomeDB: a comprehensive database and cloud-based analysis platform for translatome sequencing data. Nucleic Acids Res 46:D206–D212. https://doi.org/10.1093/nar/gkx1034
Mai Z, Xiao C, Jin J, Zhang G (2017) Low-cost, low-bias and low-input RNA-seq with High experimental verifiability based on semiconductor sequencing. Sci Rep 7:1053. https://doi.org/10.1038/s41598-017-01165-w
McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9:356–369. https://doi.org/10.1038/nrg2344
Mishmar D, Ruiz-Pesini E, Brandon M, Wallace DC (2004) Mitochondrial DNA-like sequences in the nucleus (NUMTs): insights into our African origins and the mechanism of foreign DNA integration. Hum Mutat 23:125–133. https://doi.org/10.1002/humu.10304
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. https://doi.org/10.1038/nmeth.1226
Rossier BC, Baker ME, Studer RA (2015) Epithelial sodium transport and its control by aldosterone: the story of our internal environment revisited. Physiol Rev 95:297–340. https://doi.org/10.1152/physrev.00011.2014
Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen HC, Kitts PA, Murphy TD, Pruitt KD, Thibaud-Nissen F, Albracht D, Fulton RS, Kremitzki M, Magrini V, Markovic C, McGrath S, Steinberg KM, Auger K, Chow W, Collins J, Harden G, Hubbard T, Pelan S, Simpson JT, Threadgold G, Torrance J, Wood JM, Clarke L, Koren S, Boitano M, Peluso P, Li H, Chin CS, Phillippy AM, Durbin R, Wilson RK, Flicek P, Eichler EE, Church DM (2017) Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res 27:849–864. https://doi.org/10.1101/gr.213611.116
Shapiro E, Biezuner T, Linnarsson S (2013) Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet 14:618–630. https://doi.org/10.1038/nrg3542
Shi L, Guo Y, Dong C, Huddleston J, Yang H, Han X, Fu A, Li Q, Li N, Gong S, Lintner KE, Ding Q, Wang Z, Hu J, Wang D, Wang F, Wang L, Lyon GJ, Guan Y, Shen Y, Evgrafov OV, Knowles JA, Thibaud-Nissen F, Schneider V, Yu CY, Zhou L, Eichler EE, So KF, Wang K (2016) Long-read sequencing and de novo assembly of a Chinese genome. Nat Commun 7:12065. https://doi.org/10.1038/ncomms12065
Storer CG, Pascal CE, Roberts SB, Templin WD, Seeb LW, Seeb JE (2012) Rank and order: evaluating the performance of SNPs for individual assignment in a non-model organism. PLoS One 7:e49018. https://doi.org/10.1371/journal.pone.0049018
Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J (2017) 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet 101:5–22. https://doi.org/10.1016/j.ajhg.2017.06.005
Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, Guo Y, Feng B, Li H, Lu Y, Fang X, Liang H, Du Z, Li D, Zhao Y, Hu Y, Yang Z, Zheng H, Hellmann I, Inouye M, Pool J, Yi X, Zhao J, Duan J, Zhou Y, Qin J, Ma L, Li G, Yang Z, Zhang G, Yang B, Yu C, Liang F, Li W, Li S, Li D, Ni P, Ruan J, Li Q, Zhu H, Liu D, Lu Z, Li N, Guo G, Zhang J, Ye J, Fang L, Hao Q, Chen Q, Liang Y, Su Y, San A, Ping C, Yang S, Chen F, Li L, Zhou K, Zheng H, Ren Y, Yang L, Gao Y, Yang G, Li Z, Feng X, Kristiansen K, Wong GK, Nielsen R, Durbin R, Bolund L, Zhang X, Li S, Yang H, Wang J (2008) The diploid genome sequence of an Asian individual. Nature 456:60–65. https://doi.org/10.1038/nature07484
Wang T, Cui Y, Jin J, Guo J, Wang G, Yin X, He QY, Zhang G (2013) Translating mRNAs strongly correlate to proteins in a multivariate manner and their translation ratios are phenotype specific. Nucleic Acids Res 41:4743–4754. https://doi.org/10.1093/nar/gkt178
Wu X, Xu L, Gu W, Xu Q, He QY, Sun X, Zhang G (2014) Iterative genome correction largely improves proteomic analysis of nonmodel organisms. J Proteome Res 13:2724–2734. https://doi.org/10.1021/pr500369b
Xiao CL, Mai ZB, Lian XL, Zhong JY, Jin JJ, He QY, Zhang G (2014) FANSe2: a robust and cost-efficient alignment tool for quantitative next-generation sequencing applications. PLoS One 9:e94250. https://doi.org/10.1371/journal.pone.0094250
Xiao CL, Chen Y, Xie SQ, Chen KN, Wang Y, Han Y, Luo F, Xie Z (2017) MECAT: fast mapping, error correction, and de novo assembly for single-molecule sequencing reads. Nat Methods 14:1072–1074. https://doi.org/10.1038/nmeth.4432
Yang X, Chockalingam SP, Aluru S (2013) A survey of error-correction methods for next-generation sequencing. Brief Bioinform 14:56–66. https://doi.org/10.1093/bib/bbs015
Zhang G, Fedyunin I, Kirchner S, Xiao C, Valleriani A, Ignatova Z (2012) FANSe: an accurate algorithm for quantitative mapping of large scale sequencing reads. Nucleic Acids Res 40:e83. https://doi.org/10.1093/nar/gks196
Acknowledgements
This work was supported by the Ministry of Science and Technology of China ‘National Key Research and Development Program’ (2017YFA0505001/2018YFC0910201/2018YFC0910202) and the Distinguished Young Talent Award of National High-level Personnel Program of China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mai, Z., Liu, W., Ding, W. et al. Misassembly of long reads undermines de novo-assembled ethnicity-specific genomes: validation in a Chinese Han population. Hum Genet 138, 757–769 (2019). https://doi.org/10.1007/s00439-019-02032-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-019-02032-6