Abstract
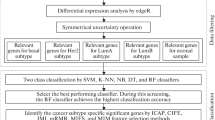
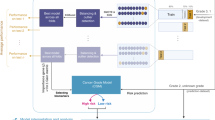
Breast cancer is the second leading cancer among women in terms of mortality rate. In recent years, its incidence frequency has been continuously rising across the globe. In this context, the new therapeutic strategies to manage the deadly disease attracts tremendous research focus. However, finding new prognostic predictors to refine the selection of therapy for the various stages of breast cancer is an unattempted issue. Aberrant expression of genes at various stages of cancer progression can be studied to identify specific genes that play a critical role in cancer staging. Moreover, while many schemes for subtype prediction in breast cancer have been explored in the literature, stage-wise classification remains a challenge. These observations motivated the proposed two-phased method: stage-specific gene signature selection and stage classification. In the first phase, meta-analysis of gene expression data is conducted to identify stage-wise biomarkers that were then used in the second phase of cancer classification. From the analysis, 118, 12 and 4 genes respectively in stage I, stage II and stage III are determined as potential biomarkers. Pathway enrichment, gene network and literature analysis validate the significance of the identified genes in breast cancer. In this study, machine learning methods were combined with principal component and posterior probability analysis. Such a scheme offers a unique opportunity to build a meaningful model for predicting breast cancer staging. Among the machine learning models compared, Support Vector Machine (SVM) is found to perform the best for the selected datasets with an accuracy of 92.21% during test data evaluation. Perhaps, biomarker identification performed here for stage-specific cancer treatment would be a meaningful step towards predictive medicine. Significantly, the determination of correct cancer stage using the proposed 134 gene signature set can possibly act as potential target for breast cancer therapeutics.






Similar content being viewed by others
Data availability
All large-scale data used in this study were obtained from online data repositories and the manuscript provide clear information on how to access the data.
References
Agarwal R, Gonzalez-Angulo AM, Myhre S, Carey M, Lee JS, Overgaard J, Alsner J, Stemke-Hale K, Lluch A, Neve RM et al (2009) Integrative analysis of cyclin protein levels identifies cyclin b1 as a classifier and predictor of outcomes in breast cancer. Clin Cancer Res 15(11):3654–3662
Aibar S, Fontanillo C, Droste C, Roson-Burgo B, Campos-Laborie FJ, Hernandez-Rivas JM, De Las Rivas J (2015) Analyse multiple disease subtypes and build associated gene networks using genome-wide expression profiles. BMC Genom 16(S5):S3
Aleskandarany MA, Vandenberghe ME, Marchiò C, Ellis IO, Sapino A, Rakha EA (2018) Tumour heterogeneity of breast cancer: from morphology to personalised medicine. Pathobiology 85(1–2):23–34
Beattie J, Hawsawi Y, Alkharobi H, El-Gendy R (2015) Igfbp-2 and- 5: important regulators of normal and neoplastic mammary gland physiology. J Cell Commun Signal 9(2):151–158
Blows FM, Driver KE, Schmidt MK, Broeks A, Van Leeuwen FE, Wesseling J, Cheang MC, Gelmon K, Nielsen TO, Blomqvist C et al (2010) Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med 7(5):e1000279
Brown MP, Grundy WN, Lin D, Cristianini N, Sugnet C, Ares M, Haussler D (1999) Support vector machine classification of microarray gene expression data. University of California, Santa Cruz
Brown MP, Grundy WN, Lin D, Cristianini N, Sugnet CW, Furey TS, Ares M, Haussler D (2000) Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc Natl Acad Sci 97(1):262–267
Castellana B, Escuin D, Peirò G, Garcia-Valdecasas B, Vázquez T, Pons C, Pérez-Olabarria M, Barnadas A, Lerma E (2012) Aspn and gjb2 are implicated in the mechanisms of invasion of ductal breast carcinomas. J Cancer 3:175
Chang CC, Lin CJ (2011) Libsvm: a ibrary for support vector machines. ACM Transact Intell Syst Technol 2(3):1–27
Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, Perou CM, Nielsen TO (2008) Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res 14(5):1368–1376
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20(3):273–297
Debnath J (2011) The multifaceted roles of autophagy in tumors-implications for breast cancer. J Mammary Gland Biol Neoplasia 16(3):173
Dettogni RS, Stur E, Laus AC, da Costa Vieira RA, Marques MMC, Santana IVV, Pulido JZ, Ribeiro LF, de Jesus Parmanhani N, Agostini LP et al (2020) Potential biomarkers of ductal carcinoma in situ progression. BMC Cancer 20(1):119
El Sayed R, El Jamal L, El Iskandarani S, Kort J, Abdelsalam M, Assi HI (2019) Endocrine and targeted therapy for hormone-receptor-positive, her2-negative advanced breast cancer: seuencing treatment and overcoming resistance. Front Oncol 9:510
Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B et al (2018) The reactome pathway knowledgebase. Nucleic Acids Res 46(D1):D649–D655
Fang M, Yuan J, Peng C, Li Y (2014) Collagen as a double-edged sword in tumor progression. Tumor Biol 35(4):2871–2882
Gamberger D, Lavrač N, Železnỳ F, Tolar J (2004) Induction of comprehensible models for gene expression datasets by subgroup discovery methodology. J Biomed Inform 37(4):269–284
Goebel C, Louden CL, McKenna R, Onugha O, Wachtel A, Long T (2019) Diagnosis of non-small cell lung cancer for early stage asymptomatic patients. Cancer Genom Proteom 16(4):229–244
Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA et al (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286(5439):531–537
Güler EN (2017) Gene expression profiling in breast cancer and its effect on therapy selection in early-stage breast cancer. Eur J Breast Health 13(4):168
Hamzeh O, Alkhateeb A, Zheng JZ, Kandalam S, Leung C, Atikukke G, Cavallo-Medved D, Palanisamy N, Rueda L (2019) A hierarchical machine learning model to discover gleason grade-specific biomarkers in prostate cancer. Diagnostics 9(4):219
Hamzeh O, Alkhateeb A, Zheng J, Kandalam S, Rueda L (2020) Prediction of tumor location in prostate cancer tissue using a machine learning system on gene expression data. BMC Bioinform 21(2):1–10
Hanna M, Diorio C (2013) Does mammographic density reflect the expression of breast cancer markers? Climacteric 16(4):407–416
Hens AB, Tiwari MK (2012) Computational time reduction for credit scoring: an integrated approach based on support vector machine and stratified sampling method. Expert Syst Appl 39(8):6774–6781
Heo KS (2019) Regulation of post-translational modification in breast cancer treatment. BMB Rep 52(2):113
Hu Z, Fan C, Oh DS, Marron J, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L et al (2006) The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genom 7(1):1–12
Jagga Z, Gupta D (2014) Classification models for clear cell renal carcinoma stage progression, based on tumor RNAseq expression trained supervised machine learning algorithms. BMC Proceedings, vol 8. Springer, Berlin, p S2
Jena MK, Janjanam J (2018) Role of extracellular matrix in breast cancer development: a brief update. F1000Research. https://doi.org/10.12688/f1000research.14133.2
Jeong SB, Im JH, Yoon JH, Bui QT, Lim SC, Song JM, Shim Y, Yun J, Hong J, Kang KW (2018) Essential role of polo-like kinase 1 (plk1) oncogene in tumor growth and metastasis of tamoxifen-resistant breast cancer. Mol Cancer Ther 17(4):825–837
Kanehisa M, Goto S (2000) Kegg: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30
Ke X, Shen L (2017) Molecular targeted therapy of cancer: The progress and future prospect. Front Lab Med 1(2):69–75
Kendziorski C, Newton M, Lan H, Gould M (2003) On parametric empirical Bayes methods for comparing multiple groups using replicated gene expression profiles. Stat Med 22(24):3899–3914
Knudsen S (2006) Cancer diagnostics with DNA microarrays. Wiley, Hoboken
Konstantinos PKS, Darlix A, Jacot W, Blom AM (2019) High levels of cartilage oligomeric matrix protein in the serum of breast cancer patients can serve as an independent prognostic marker. Front Oncol 9:11–41
Leblanc R, Peyruchaud O (2016) The role of platelets and megakaryocytes in bone metastasis. J Bone Oncol 5(3):109–111
Li F, Yang M, Li Y, Zhang M, Wang W, Yuan D, Tang D (2020) An improved clear cell renal cell carcinoma stage prediction model based on gene sets. BMC Bioinform 21:1–15
Li J, Holm J, Bergh J, Eriksson M, Darabi H, Lindström LS, Törnberg S, Hall P, Czene K (2015) Breast cancer genetic risk profile is differentially associated with interval and screen-detected breast cancers. Ann Oncol 26(3):517–522
Li X, Cowell JK, Sossey-Alaoui K (2004) Clca2 tumour suppressor gene in 1p31 is epigenetically regulated in breast cancer. Oncogene 23(7):1474–1480
Lien HC, Lee YH, Juang YL, Lu YT (2019) Fibrillin-1, a novel tgf-beta-induced factor, is preferentially expressed in metaplastic carcinoma with spindle sarcomatous metaplasia. Pathology 51(4):375–383
Tt Liu, Xs Liu, Zhang M, Xn Liu, Fx Zhu, Fm Zhu, Sw Ouyang, Sb Li, Cl Song, Hm Sun et al (2018) Cartilage oligomeric matrix protein is a prognostic factor and biomarker of colon cancer and promotes cell proliferation by activating the akt pathway. J Cancer Res Clin Oncol 144(6):1049–1063
Lun A (2020) BiocSingular: singular value decomposition for bioconductor packages. R Project for Statistical Computing, Vienna
Malvia S, Bagadi SAR, Pradhan D, Chintamani C, Bhatnagar A, Arora D, Sarin R, Saxena S (2019) Study of gene expression profiles of breast cancers in Indian women. Sci Rep 9(1):1–15
Meyer D, Dimitriadou E, Hornik K, Weingessel A, Leisch F (2019) e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071). TU Wien, Vienna
Moler E, Chow M, Mian I (2000) Analysis of molecular profile data using generative and discriminative methods. Physiol Genom 4(2):109–126
Network CGA et al (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61
Nishimura D (2001) Biocarta. Biotech Softw Internet Rep 2(3):117–120
Park JH, Katagiri T, Nakamura Y (2008) Pbk/topk, a mitotic ser/thr kinase, is a novel druggable target for breast cancer therapy. Cancer Cell Int. https://doi.org/10.1186/s12935-015-0178-0
Pavlidis P, Weston J, Cai J, Noble WS (2002) Learning gene functional classifications from multiple data types. J Comput Biol 9(2):401–411
Perou CM, Sørlie T, Eisen MB, Van De Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA et al (2000) Molecular portraits of human breast tumours. Nature 406(6797):747–752
Prydz K (2015) Determinants of glycosaminoglycan (gag) structure. Biomolecules 5(3):2003–2022
Ragunath P, Reddy BV, Abhinand P, Ahmed SS (2012) Relevance of systems biological approach in the differential diagnosis of invasive lobular carcinoma & invasive ductal carcinoma. Bioinformation 8(8):359
Ratajczak-Wielgomas K, Grzegrzolka J, Piotrowska A, Matkowski R, Wojnar A, Rys J, Ugorski M, Dziegiel P (2017) Expression of periostin in breast cancer cells. Int J Oncol 51(4):1300–1310
Roy R, Winteringham LN, Lassmann T, Forrest AR (2019) Expression levels of therapeutic targets as indicators of sensitivity to targeted therapeutics. Mol Cancer Ther 18(12):2480–2489
Saha SK, Yin Y, Chae HS, Cho SG (2019) Opposing regulation of cancer properties via krt19-mediated differential modulation of wnt/\(\beta\)-catenin/notch signaling in breast and colon cancers. Cancers 11(1):99
Saha T (2012) Lamp2a overexpression in breast tumors promotes cancer cell survival via chaperone-mediated autophagy. Autophagy 8(11):1643–1656
Sales G, Calura E, Cavalieri D, Romualdi C (2012) graphite-a bioconductor package to convert pathway topology to gene network. BMC Bioinform 13(1):20
Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH (2009) Pid: the pathway interaction database. Nucleic Acids Res 37(suppl–1):D674–D679
Sharov AA, Dudekula DB, Ko MS (2005) A web-based tool for principal component and significance analysis of microarray data. Bioinformatics 21(10):2548–2549
Singh NP, Bapi RS, Vinod P (2018) Machine learning models to predict the progression from early to late stages of papillary renal cell carcinoma. Comput Biol Med 100:92–99
Soni A, Ren Z, Hameed O, Chanda D, Morgan CJ, Siegal GP, Wei S (2015) Breast cancer subtypes predispose the site of distant metastases. Am J Clin Pathol 143(4):471–478
Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, Van De Rijn M, Jeffrey SS et al (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci 98(19):10869–10874
Sorlie T, Tibshirani R, Parker J, Hastie T, Marron J, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S et al (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100(14):8418–8423
Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET (2003) Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci 100(18):10393–10398
Tarca AL, Draghici S, Khatri P, Hassan SS, Mittal P, Js Kim, Kim CJ, Kusanovic JP, Romero R (2009) A novel signaling pathway impact analysis. Bioinformatics 25(1):75–82
Testa U, Castelli G, Pelosi E (2020) Breast cancer: a molecularly heterogenous disease needing subtype-specific treatments. Med Sci 8(1):18
Todorov H, Fournier D, Gerber S (2018) Principal components analysis: theory and application to gene expression data analysis. Genom Comput Biol 4(2):e100041–e100041
Turner NC, Neven P, Loibl S, Andre F (2017) Advances in the treatment of advanced oestrogen-receptor-positive breast cancer. Lancet 389(10087):2403–2414
Vallejos CS, Gómez HL, Cruz WR, Pinto JA, Dyer RR, Velarde R, Suazo JF, Neciosup SP, León M, Miguel A et al (2010) Breast cancer classification according to immunohistochemistry markers: subtypes and association with clinicopathologic variables in a peruvian hospital database. Clin Breast Cancer 10(4):294–300
Vendrell J, Magnino F, Danis E, Duchesne M, Pinloche S, Pons M, Birnbaum D, Nguyen C, Theillet C, Cohen P (2004) Estrogen regulation in human breast cancer cells of new downstream gene targets involved in estrogen metabolism, cell proliferation and cell transformation. J Mol Endocrinol 32(2):397–414
Villman K, Sjöström J, Heikkilä R, Hultborn R, Malmström P, Bengtsson NO, Söderberg M, Saksela E, Blomqvist C (2006) Top2a and her2 gene amplification as predictors of response to anthracycline treatment in breast cancer. Acta Oncol 45(5):590–596
Wall ME, Rechtsteiner A, Rocha LM (2003) Singular value decomposition and principal component analysis. A practical approach to microarray data analysis. Springer, Berlin, pp 91–109
Wang S, Wang J, Chen H, Zhang B (2006) Svm-based tumor classification with gene expression data. International conference on advanced data mining and applications. Springer, Berlin, pp 864–870
Weigel MT, Dowsett M (2010) Current and emerging biomarkers in breast cancer: prognosis and prediction. Endocr Relat Cancer 17(4):R245–R262
WHO (2020) WHO report on cancer: setting priorities, investing wisely and providing care for all. World Health Organization, Geneva
Wirapati P, Sotiriou C, Kunkel S, Farmer P, Pradervand S, Haibe-Kains B, Desmedt C, Ignatiadis M, Sengstag T, Schütz F et al (2008) Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res 10(4):R65
Wonsey DR, Follettie MT (2005) Loss of the forkhead transcription factor foxm1 causes centrosome amplification and mitotic catastrophe. Can Res 65(12):5181–5189
Wu MZ, Chen SF, Nieh S, Benner C, Ger LP, Jan CI, Ma L, Chen CH, Hishida T, Chang HT et al (2015) Hypoxia drives breast tumor malignancy through a tet-\(\text{ tnf }\alpha\)-p38-mapk signaling axis. Can Res 75(18):3912–3924
Wu Y, Wu P, Zhang Q, Chen W, Liu X, Zheng W (2019) Mfap5 promotes basal-like breast cancer progression by activating the EMT program. Cell Biosci 9(1):24
Yang Y, Li DP, Shen N, Yu XC, Li JB, Song Q, Zhang JH (2015) Tpx2 promotes migration and invasion of human breast cancer cells. Asian Pac J Trop Med 8(12):1064–1070
Yeom YK, Chae EY, Kim HH, Cha JH, Shin HJ, Choi WJ (2019) Screening mammography for second breast cancers in women with history of early-stage breast cancer: factors and causes associated with non-detection. BMC Med Imaging 19(1):1–9
Yi T, Zhou X, Sang K, Huang X, Zhou J, Ge L (2019) Activation of lncrna lnc-slc4a1-1 induced by h3k27 acetylation promotes the development of breast cancer via activating cxcl8 and nf-kb pathway. Artif Cells Nanomed Biotechnol 47(1):3765–3773
Yip GW, Smollich M, Götte M (2006) Therapeutic value of glycosaminoglycans in cancer. Mol Cancer Ther 5(9):2139–2148
Yuan M, Newton M, Sarkar D, Kendziorski C (2017) Ebarrays: unified approach for simultaneous gene clustering and differential expression identification. Biometrics. https://doi.org/10.1111/j.1541-0420.2006.00611.x
Zhou Y, Rucker EB III, Zhou BP (2016) Autophagy regulation in the development and treatment of breast cancer. Acta Biochim Biophys Sin 48(1):60–74
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest, financial or otherwise.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors
Additional information
Communicated by Shuhua Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Athira, K., Gopakumar, G. Breast cancer stage prediction: a computational approach guided by transcriptome analysis. Mol Genet Genomics 297, 1467–1479 (2022). https://doi.org/10.1007/s00438-022-01932-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-022-01932-z