Abstract
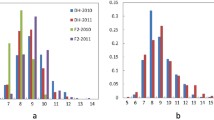
Increasing the yield of rapeseed is required to meet the rapidly expanding demand for both edible vegetable oil and biofuel. Branching, an important determinant of yield potential in rapeseed, is controlled by a series of quantitative trait loci (QTLs). To explore the genetic mechanism regulating the natural variation of branching, a BC1F1 population derived from a cross between dense branching 2 (dense branching line) and L72 (normal branching line) was used to map QTL conferring branching in rapeseed. A major QTL, qDB.A03, for branching-related traits was identified by the BeadChip Array assisted bulked segregation analysis method, which was subsequently validated by the classical QTL mapping approach, and fine mapped to the 256 kb region. This interval contains 56 annotated or predicted genes, 8 of which are candidates for controlling the branching trait. Comparative and expression analysis revealed four promising candidate genes for qDB.A03. Fine mapping and identification of the candidate genes for qDB.A03 represents the first step toward unraveling the genetical and molecular mechanisms controlling branching in rapeseed.






Similar content being viewed by others
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30:174–178
Betsuyaku S, Takahashi F, Kinoshita A, Miwa H, Shinozaki K, Fukuda H, Sawa S (2011) Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol 52:14–29
Chen W, Zhang Y, Liu X, Chen B, Tu J, Tingdong F (2007) Detection of QTL for six yield-related traits in oilseed rape (Brassica napus) using DH and immortalized F2 populations. Theor Appl Genet 115:849–858
Chen B, Xu K, Li J, Li F, Qiao J, Li H, Gao G, Yan G, Wu X (2014) Evaluation of yield and agronomic traits and their genetic variation in 488 global collections of Brassica napus L. Genet Resour Crop Ev 61:979–999
Clarke JM, Simpson GM (1978) Influence of irrigation and seeding rates on yield and yield component. Can J Plant Sci 58:731–737
Clarke WE, Higgins EE, Plieske J, Wieseke R, Sidebottom C, Khedikar Y, Batley J, Edwards D, Meng J, Li R, Lawley CT, Pauquet J, Laga B, Cheung W, Iniguez-Luy F, Dyrszka E, Rae S, Stich B, Snowdon RJ, Sharpe AG, Ganal MW, Parkin IA (2016) A high-density SNP genotyping array for Brassica napus and its ancestral diploid species based on optimised selection of single-locus markers in the allotetraploid genome. Theor Appl Genet 129:1887–1899
Di Giacomo E, Serino G, Frugis G (2013) Emerging role of the ubiquitin proteasome system in the control of shoot apical meristem function(f). J Integr Plant Biol 55:7–20
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Fu T, Yang G, Tu J (2001) The present and future of rapeseed production in China. In: Proceedings of International Symposium on Rapeseed Science. Science Press Ltd, NewYork, pp 3–5
Girin T, Paicu T, Stephenson P, Fuentes S, Korner E, O’Brien M, Sorefan K, Wood TA, Balanza V, Ferrandiz C, Smyth DR, Ostergaard L (2011) INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell 23:3641–3653
He Y, Wu D, Wei D, Fu Y, Cui Y, Dong H, Tan C, Qian W (2017) GWAS, QTL mapping and gene expression analyses in Brassica napus reveal genetic control of branching morphogenesis. Sci Rep 7
Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15:1899–1911
Hu Q, Hua W, Yin Y, Zhang X, Liu L, Shi J, Zhao Y, Qin L, Chen C, Wang H (2017) Rapeseed research and production in China. Crop J 5:127–135
Hu L, Zhang H, Sun Y, Shen X, Amoo O, Wang Y, Fan C, Zhou Y (2020) BnA10.RCO, a homeobox gene, positively regulates leaf lobe formation in Brassica napus L. Theor Appl Genet 133:3333–3343
Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41:494–497
Jaganathan D, Bohra A, Thudi M, Varshney RK (2020) Fine mapping and gene cloning in the post-NGS era: advances and prospects. Theor Appl Genet 133:1791–1810
Jia H, Li M, Li W, Liu L, Jian Y, Yang Z, Shen X, Ning Q, Du Y, Zhao R, Jackson D, Yang X, Zhang Z (2020) A serine/threonine protein kinase encoding gene KERNEL NUMBER PER ROW6 regulates maize grain yield. Nat Commun 11(1):988
Khan SU, Saeed S, Khan MHU, Fan C, Ahmar S, Arriagada O, Shahzad R, Branca F, Mora-Poblete F (2021) Advances and Challenges for QTL Analysis and GWAS in the Plant-Breeding of High-Yielding: A Focus on Rapeseed. Biomolecules 11
Kitagawa M, Jackson D (2019) Control of Meristem Size. Annu Rev Plant Biol 70:269–291
Kuhn A, Runciman B, Tasker-Brown W, Ostergaard L (2019) Two Auxin Response Elements Fine-Tune PINOID Expression During Gynoecium Development in Arabidopsis thaliana. Biomolecules 9
Laux T, Mayer K, Berger J, Jurgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122:87–96
Lee H, Jun YS, Cha OK, Sheen J (2019) Mitogen-activated protein kinases MPK3 and MPK6 are required for stem cell maintenance in the Arabidopsis shoot apical meristem. Plant Cell Rep 38:311–319
Li D, Fu X, Guo L, Huang Z, Li Y, Liu Y, He Z, Cao X, Ma X, Zhao M, Zhu G, Xiao L, Wang H, Chen X, Liu R, Liu X (2016a) FAR-RED ELONGATED HYPOCOTYL3 activates SEPALLATA2 but inhibits CLAVATA3 to regulate meristem determinacy and maintenance in Arabidopsis. Proc Natl Acad Sci U S A 113:9375–9380
Li F, Chen B, Xu K, Gao G, Yan G, Qiao J, Li J, Li H, Li L, Xiao X, Zhang T, Nishio T, Wu X (2016b) A genome-wide association study of plant height and primary branch number in rapeseed (Brassica napus). Plant Sci 242:169–177
Li S, Zhu Y, Varshney RK, Zhan J, Zheng X, Shi J, Wang X, Liu G, Wang H (2019) A systematic dissection of the mechanisms underlying the natural variation of silique number in rapeseed (Brassica napus L.) germplasm. Plant Biotechnol J 18:568–580
Li B, Gao J, Chen J, Wang Z, Shen W, Yi B, Wen J, Ma C, Shen J, Fu T, Tu J (2020) Identification and fine mapping of a major locus controlling branching in Brassica napus. Theor Appl Genet 133:771–783
Lincoln S (1992a) Construcing genetic maps with MAPMAKER/EXP 3.0. Whitehead Institute Technical Report
Lincoln S (1992b) Mapping genes controlling quantitative traits with MAPMAKER/QTL 1.1. Whitehead institute technical report
Luo H, Pandey MK, Khan AW, Guo J, Wu B, Cai Y, Huang L, Zhou X, Chen Y, Chen W, Liu N, Lei Y, Liao B, Varshney RK, Jiang H (2019) Discovery of genomic regions and candidate genes controlling shelling percentage using QTL-seq approach in cultivated peanut (Arachis hypogaea L.). Plant Biotechnol J 17:1248–1260
Luo X, Ma C, Yue Y, Hu K, Li Y, Duan Z, Wu M, Tu J, Shen J, Yi B, Fu T (2015) Unravelling the complex trait of harvest index in rapeseed (Brassica napus L.) with association mapping. BMC Genomics 16
Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95:805–815
McSteen P, Leyser O (2005) Shoot Branching. Annu Rev Plant Biol 56:353–374
Moubayidin L, Ostergaard L (2014) Dynamic control of auxin distribution imposes a bilateral-to-radial symmetry switch during gynoecium development. Curr Biol 24:2743–2748
Petrovska B, Cenklova V, Pochylova Z, Kourova H, Doskocilova A, Plihal O, Binarova L, Binarova P (2012) Plant Aurora kinases play a role in maintenance of primary meristems and control of endoreduplication. New Phytol 193:590–604
Rodriguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB (2017) Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 171:470–480 e478
Sahu PK, Sao R, Mondal S, Vishwakarma G, Gupta SK, Kumar V, Singh S, Sharma D, Das BK (2020) Next Generation Sequencing Based Forward Genetic Approaches for Identification and Mapping of Causal Mutations in Crop Plants: A Comprehensive Review. Plants (Basel) 9
Shen W, Qin P, Yan M, Li B, Wu Z, Wen J, Yi B, Ma C, Shen J, Fu T, Tu J (2019) Fine mapping of a silique length- and seed weight-related gene in Brassica napus. Theor Appl Genet 132:2985–2996
Shi J, Li R, Zou J, Long Y, Meng J (2011) A dynamic and complex network regulates the heterosis of yield-correlated traits in rapeseed (Brassica napus L.). PLoS One 6:e21645
Singh VK, Khan AW, Jaganathan D, Thudi M, Roorkiwal M, Takagi H, Garg V, Kumar V, Chitikineni A, Gaur PM, Sutton T, Terauchi R, Varshney RK (2016) QTL-seq for rapid identification of candidate genes for 100-seed weight and root/total plant dry weight ratio under rainfed conditions in chickpea. Plant Biotechnol J 14:2110–2119
Somssich M, Je BI, Simon R, Jackson D (2016) CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143:3238–3248
Sonoda Y, Yao SG, Sako K, Sato T, Kato W, Ohto MA, Ichikawa T, Matsui M, Yamaguchi J, Ikeda A (2007) SHA1, a novel RING finger protein, functions in shoot apical meristem maintenance in Arabidopsis. Plant J 50:586–596
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, Innan H, Cano LM, Kamoun S, Terauchi R (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74:174–183
Tester M, Langridge P (2010) Breeding Technologies to Increase Crop Production in a Changing World. Science 327:818–822
Tunçtürk M, Çiftçi V (2007) Relationships between yield and some yi̇eld components in rapeseed (Brassica napus ssp. oleifera L.) cultivars by usi̇ng correlation and path analysis. Pak J of Bot 39:81–84
Valluru M, Sorefan K (2020) Control of stem cell niche and fruit development in Arabidopsis thaliana by AGO10/ZWL requires the bHLH transcription factor INDEHISCENT. bioRxiv
Wang Y, Jiao Y (2018) Axillary meristem initiation — a way to branch out. Curr Opin Plant Biol 41:61–66
Wang B, Smith SM, Li J (2018) Genetic Regulation of Shoot Architecture. Annu Rev Plant Biol 69:437–468
Wei C, Zhu L, Wen J, Yi B, Ma C, Tu J, Shen J, Fu T (2018) Morphological, transcriptomics and biochemical characterization of new dwarf mutant of Brassica napus. Plant Sci 270:97–113
Williams L, Fletcher JC (2005) Stem cell regulation in the Arabidopsis shoot apical meristem. Curr Opin Plant Biol 8:582–586
Xiao L, Yi B, Chen Y, Huang Z, Chen W, Ma C, Tu J, Fu T (2008) Molecular markers linked to Bn;rf: a recessive epistatic inhibitor gene of recessive genic male sterility in Brassica napus L. Euphytica 164:377–384
Xiao Q, Wang H, Song N, Yu Z, Imran K, Xie W, Qiu S, Zhou F, Wen J, Dai C, Ma C, Tu J, Shen J, Fu T, Yi B (2021) The Bnapus50K array: a quick and versatile genotyping tool for Brassica napus genomic breeding and research. G3 (Bethesda) 11
Xing S, Rosso MG, Zachgo S (2005) ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132:1555–1565
Xu J, Song X, Cheng Y, Zou X, Zeng L, Qiao X, Lu G, Fu G, Qu Z, Zhang X (2014) Identification of QTLs for Branch Number in Oilseed Rape (Brassica napus L.). J Genet Genomics 41:557-559
Xue Z, Liu L, Zhang C (2020) Regulation of Shoot Apical Meristem and Axillary Meristem Development in Plants. Int J Mol Sci 21
Yang M, Jiao Y (2016) Regulation of axillary meristem initiation by transcription factors and plant hormones. Front Plant Sci 7:183
Yin XJ, Volk S, Ljung K, Mehlmer N, Dolezal K, Ditengou F, Hanano S, Davis SJ, Schmelzer E, Sandberg G, Teige M, Palme K, Pickart C, Bachmair A (2007) Ubiquitin lysine 63 chain forming ligases regulate apical dominance in Arabidopsis. Plant Cell 19:1898–1911
Zhang Y, Wang T, Guo Y, Liu X, Li B, Li M (2021) Genetic Diversity of Core Collection of Oilseed Rape from Hunan Province Revealed by SNP Chips. Mol Plant Breed 1:10
Zhang Y, Zhang D, Yu H, Lin B, Fu Y, Hua S (2016) Floral Initiation in Response to Planting Date Reveals the Key Role of Floral Meristem Differentiation Prior to Budding in Canola (Brassica napus L.). Front Plant Sci 7:1369
Zhao W, Wang X, Wang H, Tian J, Li B, Chen L, Chao H, Long Y, Xiang J, Gan J, Liang W, Li M (2016) Genome-wide identification of QTL for seed yield and yield-related traits and construction of a high-density consensus map for QTL comparison in Brassica napus. Front Plant Sci 7:17
Zheng M, Peng C, Liu H, Tang M, Yang H, Li X, Liu J, Sun X, Wang X, Xu J, Hua W, Wang H (2017) Genome-Wide Association Study Reveals Candidate Genes for Control of Plant Height, Branch Initiation Height and Branch Number in Rapeseed (Brassica napus L.). Front Plant Sci 8
Zhu Y, Wagner D (2020) Plant Inflorescence Architecture: The Formation, Activity, and Fate of Axillary Meristems. Cold Spring Harb Perspect Biol 12:034652
Acknowledgements
This work was financed by the Agricultural Science and Technology Innovation Program of Hunan (2020CX28), the Key Research and Development Program in Hunan province (2020NK2045), the National Key Research and Development Program of China (2016YFD010300).
Author information
Authors and Affiliations
Contributions
BL performed research and wrote the paper. WT and YG helped with the experiments. XL, LD and LQ gave advises to the experimental design. ML conceived the project and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical standards
The authors declare that this study complies with current laws of China.
Additional information
Communicated by Bing Yang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, B., Wang, T., Guo, Y. et al. Fine mapping of qDB.A03, a QTL for rapeseed branching, and identification of the candidate gene. Mol Genet Genomics 297, 699–710 (2022). https://doi.org/10.1007/s00438-022-01881-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-022-01881-7