Abstract
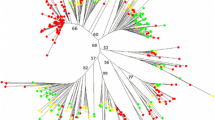
Transposable elements (TEs) are the most abundant genetic material for almost all eukaryotic genomes. Their effects on the host genomes range from an extensive size variation to the regulation of gene expression, altering gene function and creating new genes. Because of TEs pivotal contribute to the host genome structure and regulation, their identification and characterization provide a wealth of useful data for gaining an in-depth understanding of host genome functioning. The giant reed (Arundo donax) is a perennial rhizomatous C3 grass, octadecaploid, with an estimated nuclear genome size of 2744 Mbp. It is a promising feedstock for second-generation biofuels and biomethane production. To identify and characterize the most repetitive TEs in the genomes of A. donax and its ancestral A. plinii species, we carried out low-coverage whole genome shotgun sequencing for both species. Using a de novo repeat identification approach, 33,041 and 28,237 non-redundant repetitive sequences were identified and characterized in A. donax and A. plinii genomes, representing 37.55 and 31.68% of each genome, respectively. Comparative phylogenetic analyses, including the major TE classes identified in A. donax and A. plinii, together with rice and maize TE paralogs, were carried out to understand the evolutionary relationship of the most abundant TE classes. Highly conserved copies of RIRE1-like Ty1-Copia elements were discovered in two Arundo spp. in which they represented nearly 3% of each genomic sequence. We identified and characterized the medium/highly repetitive TEs in two unexplored polyploid genomes, thus generating useful information for the study of the genomic structure, composition, and functioning of these two non-model species. We provided a valuable resource that could be exploited in any effort aimed at sequencing and assembling these two genomes.






Similar content being viewed by others
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bailly-Bechet M, Haudry A, Lerat E (2014) “One code to find them all”: a perl tool to conveniently parse RepeatMasker output files. Mob DNA 5:13
Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, Westerman RP, SanMiguel PJ, Bennetzen JL (2009) Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. doi:10.1371/journal.pgen.1000732
Bennett MD, Leitch IJ (2004) Angiosperm DNA C-values database (release 5.0, Dec. 2004). http://www.kew.org/cvalues/homepage.html. Accessed 1 July 2016
Bennetzen JL (2005) Transposable elements, gene creation and genome rearrangement in flowering plants. Curr Opin Genet Dev 15:621–627
Bennetzen JL, Wang H (2014) The contributions of transposable elements to the structure, function, and evolution of plant genomes. Ann Rev Plant Biol 65:505–530
Bucci A, Cassani E, Landoni M, Cantaluppi E, Pilu R (2013) Analysis of chromosome number and speculations on the origin of Arundo donax L. (Giant Reed). Cytol Genet 47:237–241
Claros MG, Bautista R, Guerrero-Fernández D, Benzerki H, Seoane P, Fernández-Pozo N (2012) Why assembling plant genome sequences is so challenging. Biology (Basel) 1:439–459
de Setta N, Monteiro-Vitorello CB, Metcalfe CJ, Cruz GM, Del Bem LE, Vicentini R, Nogueira FT, Campos RA, Nunes SL, Turrini PC, Vieira AP, Ochoa Cruz EA, Corrêa TC, Hotta CT, de Mello Varani A, Vautrin S, da Trindade AS, de Mendonça Vilela M, Lembke CG, Sato PM, de Andrade RF, Nishiyama MY Jr, Cardoso-Silva CB, Scortecci KC, Garcia AA, Carneiro MS, Kim C, Paterson AH, Bergès H, D’Hont A, de Souza AP, Souza GM, Vincentz M, Kitajima JP, Van Sluys MA (2014) Building the sugarcane genome for biotechnology and identifying evolutionary trends. BMC Genom 15:540
Devos KM, Brown JKM, Bennetzen JL (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res 12:1075–1079
Domingues DS, Cruz GM, Metcalfe CJ, Nogueira FT, Vicentini R, de Salves C, Van Sluys MA (2012) Analysis of plant LTR-retrotransposons at the fine-scale family level reveals individual molecular patterns. BMC Genom 13:137
e Silva CFL, Schirmer MA, Maeda RN, Barcelos CA, Pereira N Jr (2015) Potential of giant reed (Arundo donax L.) for second generation ethanol production. Electron J Biotechnol 18:10–15
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004(32):1792–1797
El Baidouri M, Carpentier MC, Cooke R, Gao D, Lasserre E, Llauro C, Mirouze M, Picault N, Jackson SA, Panaud O (2014) Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res 24:831–838
Feschotte C, Pritham EJ (2007) DNA transposons and the evolution of eukaryotic genomes. Ann Rev Genet 41:331–368
Feschotte C, Jiang N, Wessler SR (2002) Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3:329–341
Finnegan DJ (1989) Eukaryotic transposable elements and genome evolution. Trends Genet 5:103–107
Gaut BS, Ross-Ibarra J (2008) Selection on major components of angiosperm genomes. Science 320:484–486
Gebre YG, Bertolini E, Pè ME, Zuccolo A (2016) Identification and characterizationof abundant repetitive sequences in Eragrostis tef cv. Enatite genome. BMC Plant Biol 16:39
Hardion L, Verlaque R, Callmander MW, Vila B (2012a) Arundo micrantha Lam. (Poaceae), the correct name for Arundo mauritanica Desf. and Arundo mediterranea Danin. Candollea 67:131–135
Hardion L, Verlaque R, Rbaumel A, Juin M, Vila B (2012b) Revised systematics of Mediterranean Arundo (Poaceae) based on AFLP fingerprints and morphology. Taxon 61:1217–1226
Hardion L, Verlaque R, Saltonstall K, Leriche A, Vila B (2014) Origin of the invasive Arundo donax (Poaceae): a trans-Asian expedition in herbaria. Ann Bot 114(3):455–462
Hardion L, Verlaque R, Rosato M, Rosselló JA, Vila B (2015) Impact of polyploidy on fertility variation of Mediterranean Arundo L. (Poaceae). Mol Biol Genet 338:298–306
Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF (2006) Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res 16(10):1252–1261
Hollister JD, Gaut BS (2009) Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res 19:1419–1428
Huang X, Madan A (1999) CAP 3: a DNA sequence assembly program. Genome Res 9:868–877
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J (2005) Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110:462–467
Jurka J, Bao W, Kojima K, Kapitonov VV (2011) Repetitive elements: bioinformatic identification, classification and analysis. In: Encyclopedia of life sciences (ELS). Wiley, Chichester. doi:10.1002/9780470015902.a0005270.pub2
Komolwanich T, Tatijarern P, Prasertwasu S, Khumsupan D, Chaisuwan T, Luengnaruemitchai A, Wongkasemjit S (2014) Comparative potentiality of Kans grass (Saccharum spontaneum) and Giant reed (Arundo donax) as lignocellulosic feedstocks for the release of monomeric sugars by microwave/chemical pretreatment. Cellulose 21:1327–1340
Kumar A, Bennetzen JL (1999) Plant retrotransposons. Ann Rev Genet 33:479–532
Lee SI, Kim NS (2014) Transposable elements and genome size variations in plants. Genomics Inform 12:87–97
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659
Ma J, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14:860–869
Macas J, Neumann P, Navrátilová A (2007) Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genom 8:427
Macas J, Kejnovský E, Neumann P, Novák P, Koblížková A, Vyskot B (2011) Next generation sequencing-based analysis of repetitive DNA in the model dioceous plant silene latifolia. PLoS One 6:e27335
Marchler-Bauer A et al (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43(Database issue):D222–D226
Mariani C, Cabrini R, Danin A, Piffanelli P, Fricano A, Gomarasca S, Dicandilo M (2010) Origin, diffusion and reproduction of the giant reed (Arundo donax L.): a promising weedy energy crop. Ann Appl Biol 157(2):191–202
McClintock B (1984) The significance of responses of the genome to challenge. Science 226:792–801
Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S et al (2015) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 43(Database issue):D213–D221
Moisy C, Schulman AH, Kalendar R, Buchmann JP, Pelsy F (2014) The Tvv1 retrotransposon family is conserved between plant genomes separated by over 100 million years. Theor Appl Genet 127:1223–1235
Oliver KR, McComb JA, Greene WK (2013) Transposable elements: powerful contributors to angiosperm evolution and diversity. Genome Biol Evol 5:1886–1901
Parisod C, Alix K, Just J, Petit M, Sarilar V, Mhiri C, Ainouche M, Chalhoub B, Grandbastien MA (2010) Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol 186:37–45
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Mehboob-ur-Rahman Ware D, Westhoff P, Mayer KF, Messing J, Rokhsar DS (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Piegu B, Buyot R, Picault A, Saniyal A, Kim HR, Collura K, Brar DS, Jackson SA, Wing RA, Panaud O (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16:1262–1269
Pilu R, Cassani E, Landoni M, Badone FC, Passera A, Cantaluppi E, Corno L, Adani F (2014) Genetic characterization of an Italian Giant Reed (Arundo donax L.) clones collection: exploiting clonal selection. Euphytica 196:169–181
Poptsova MS, Il’icheva IA, Nechipurenko DY, Panchenko LA, Khodikov MV, Oparina NY, Polozov RV, Nechipurenko YD, Grokhovsky SL (2014) Non-random DNA fragmentation in next-generation sequencing. Sci Rep. 31(4):4532
Price AL, Jones NC, Pevzner PA (2005) De novo identification of repeat families in large genomes. Bioinformatics 21:351–358
Ragaglini G, Dragoni F, Simone M, Bonari E (2014) Bioresource technology suitability of giant reed (Arundo donax L.) for anaerobic digestion: effect of harvest time and frequency on the biomethane yield potential. Bioresour Technol 152:107–115
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du F, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen W, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He R, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin J, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan C, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren L, Wei S, Kumari S, Faga B, Levy MJ, McMahan L, Van Buren P, Vaughn MV, Ying K, Yeh C, Emrich SJ, Jia Y, Kalyanaraman A, Hsia A, Barbazuk WB, Baucom RS, Brutnell TP, Carpita NC, Chaparro C, Chia J, Deragon J, Estill JC, Fu Y, Jeddeloh JA, Han Y, Lee H, Li P, Lisch DR, Liu S, Liu Z, Nagel DH, McCann MC, SanMiguel P, Myers AM, Nettleton D, Nguyen J, Penning BW, Ponnala L, Schneider KL, Schwartz DC, Sharma A, Soderlund C, Springer NM, Sun Q, Wang H, Waterman M, Westerman R, Wolfgruber TK, Yang L, Yu Y, Zhang L, Zhou S, Zhu Q, Bennetzen JL, Dawe RK, Jiang J, Jiang N, Presting GG, Wessler SR, Aluru S, Martienssen RA, Clifton SW, McCombie WR, Wing RA, Wilson RK (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Senerchia N, Felber F, Parisod C (2014) Contrasting evolutionary trajectories of multiple retrotransposons following independent allopolyploidy in wild wheats. New Phytol 202:975–985
Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8:272–285
Smit A, Hubley R, Green P (1996) RepeatMasker Open-3.0. RepeatMasker Open-3.0. http://www.repeatmasker.org. Accessed 1 July 2016
Smýkal P, Kalendar R, Ford R, Macas J, Griga M (2009) Evolutionary conserved lineage of Angela-family retrotransposons as a genome-wide microsatellite repeat dispersal agent. Heredity (Edinb) 103:157–167
Sonnhammer EL, Durbin R (1995) A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167:GC1–GC10
Spannagl M, Noubibou O, Haase D, Yang L, Gundlach H, Hindemitt T, Klee K, Haberer G, Schoof H, Mayer KF (2007) MIPSPlantsDB—plant database resource for integrative and comparative plant genome research. Nucleic Acids Res 35:D834–D840
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tanaka T, Antonio BA, Kikuchi S, Matsumoto T, Nagamura Y, Numa H, Sakai H, Wu J, Itoh T, Sasaki T, Aono R, Fujii Y, Habara T, Harada E, Kanno M, Kawahara Y, Kawashima H, Kubooka H, Matsuya A, Nakaoka H, Saichi N, Sanbonmatsu R, Sato Y, Shinso Y, Suzuki M, Takeda J, Tanino M, Todokoro F, Yamaguchi K, Yamamoto N, Yamasaki C, Imanishi T, Okido T, Tada M, Ikeo K, Tateno Y, Gojobori T, Lin YC, Wei FJ, Hsing YI, Zhao Q, Han B, Kramer MR, McCombie RW, Lonsdale D, O’Donovan CC, Whitfield EJ, Apweiler R, Koyanagi KO, Khurana JP, Raghuvanshi S, Singh NK, Tyagi AK, Haberer G, Fujisawa M, Hosokawa S, Ito Y, Ikawa H, Shibata M, Yamamoto M, Bruskiewich RM, Hoen DR, Bureau TE, Namiki N, Ohyanagi H, Sakai Y, Nobushima S, Sakata K, Barrero RA, Sato Y, Souvorov A, Smith-White B, Tatusova T, An S, An G, OOta S, Fuks G, Fuks G, Messing J, Christie KR, Lieberherr D, Kim H, Zuccolo A, Wing RA, Nobuta K, Green PJ, Lu C, Meyers BC, Chaparro C, Piegu B, Panaud O, Echeverria M (2008) The rice annotation project database (RAP-DB): 2008 update. Nucleic Acids Res 36:D1028–D1033
The UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res 43:D204–D212
Tóth G, Deák G, Barta E, Kiss GB (2006) PLOTREP: a web tool for defragmentation and visual analysis of dispersed genomic repeats. Nucleic Acids Res 34:W708–W713
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Wicker T, Narechania A, Sabot F, Stein J, Vu GTH, Graner A, Ware D, Stein N (2008) Low-pass shotgun sequencing of the barley genome facilitates rapid identification of genes, conserved non-coding sequences and novel repeats. BMC Genom 9:518
Xiong Y, Eickbush TH (1990) Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9:3353–3362
Zhang J, Kobert K, Flouri T, Stamatakis A (2014) PEAR: a fast and accurate illumina paired-end reAd mergeR. Bioinformatics 30:614–620
Zuccolo A, Sebastian A, Talag J, Yu Y, Kim H, Collura K, Kudrna D, Wing RA (2007) Transposable element distribution, abundance and role in genome size variation in the genus Oryza. BMC Evol Biol 7:152
Zuccolo A, Scofield DG, De Paoli E, Morgante M (2015) The Ty1-copia LTR retroelement family PARTC is highly conserved in conifers over 200 MY of evolution. Gene 568(1):89–99. doi:10.1016/j.gene.2015.05.028
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Data availability
The raw sequence data used to analyze Arundo spp. were submitted to GenBank under the BioSample Accession Numbers SAMN05853577 and SAMN05853578. Repeat libraries and sequence alignments supporting the conclusions of this research are listed in the “Additional Files” section.
Funding
This project was funded by the Scuola Superiore Sant’Anna, Pisa, Italy (APOMIS11AZ) and by the Doctoral School of Life Sciences of the Scuola Superiore Sant’Anna, Pisa, Italy. We thank Dr. Roberto Pilu from Milan University, Italy, for kindly providing A. donax and A. plinii fresh leaf tissue.
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
438_2016_1263_MOESM5_ESM.png
Dot plot comparisons of three A. donax repetitive contigs with (a) RIRE1 (complete element) (b) Copia-16_SB (internal region) (c) RLG_sclvana_6_1 (internal region) elements at nucleotide level using Dotter. A copy of 5’ has been added to the 3’ LTR of contig 19571 (PNG 102 kb)
438_2016_1263_MOESM7_ESM.meg
Copia_DPMR.meg: multiple alignment of Ty1-Copia RT paralogous sequences identified in Arundo spp., Rice and Maize (MEG 112 kb)
438_2016_1263_MOESM8_ESM.meg
Gypsy_DPMR.meg: multiple alignment of Ty3-Gypsy RT paralogous sequences identified in Arundo spp., Rice and Maize (MEG 161 kb)
438_2016_1263_MOESM9_ESM.pdf
Ty1-Copia-detailed.pdf: NJ tree of Ty1-Copia RT paralogous sequences identified in Arundo spp., Rice and Maize. It shows sequence names (PDF 238 kb)
438_2016_1263_MOESM10_ESM.pdf
Ty3-Gypsy-detailed.pdf: NJ tree of Ty3-Gypsy RT paralogous sequences identified in Arundo spp., Rice and Maize. It shows sequence names (PDF 439 kb)
438_2016_1263_MOESM13_ESM.fa
TE_tracts.fa: multifasta file including the tracts of TE coding domains used as queries in similarity searches to retrieve copies of TE paralogous (FA 1 kb)
438_2016_1263_MOESM14_ESM.pdf
COPIA_ctg.pdf: NJ tree of Ty1-Copia RT paralogous sequences identified in extended repetitive libraries for both Arundo species (PDF 81 kb)
438_2016_1263_MOESM15_ESM.pdf
GYPSY_ctg.pdf: NJ tree of Ty3-gypsy RT paralogous sequences identified in extended repetitive libraries for both Arundo species (PDF 138 kb)
438_2016_1263_MOESM16_ESM.pdf
CACTA_ctg.pdf: NJ tree of CACTA transposase paralogous sequences identified in extended repetitive libraries for both Arundo species (PDF 20 kb)
438_2016_1263_MOESM17_ESM.pdf
MUDR_ctg.pdf: NJ tree of MuDR transposase paralogous sequences identified in extended repetitive libraries for both Arundo species (PDF 9 kb)
Rights and permissions
About this article
Cite this article
Lwin, A.K., Bertolini, E., Pè, M.E. et al. Genomic skimming for identification of medium/highly abundant transposable elements in Arundo donax and Arundo plinii . Mol Genet Genomics 292, 157–171 (2017). https://doi.org/10.1007/s00438-016-1263-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-016-1263-3