Abstract
Background
This study aims to establish a predictive model for assessing the risk of esophageal cancer lung metastasis using machine learning techniques.
Methods
Data on esophageal cancer patients from 2010 to 2020 were extracted from the surveillance, epidemiology, and end results (SEER) database. Through univariate and multivariate logistic regression analyses, eight indicators related to the risk of lung metastasis were selected. These indicators were incorporated into six machine learning classifiers to develop corresponding predictive models. The performance of these models was evaluated and compared using metrics such as The area under curve (AUC), accuracy, sensitivity, specificity, and F1 score.
Results
A total of 20,249 confirmed cases of esophageal cancer were included in this study. Among them, 14,174 cases (70%) were assigned to the training set while 6075 cases (30%) constituted the internal test set. Primary site location, tumor histology, tumor grade classification system T staging criteria N staging criteria brain metastasis bone metastasis liver metastasis emerged as independent risk factors for esophageal cancer with lung metastasis. Amongst the six constructed models, the GBM algorithm-based machine learning model demonstrated superior performance during internal dataset validation. AUC, accuracy, sensitivity, and specificity values achieved by this model stood at respectively at 0.803, 0.849, 0.604, and 0.867.
Conclusion
We have developed an online calculator based on the GBM model (https://lvgrkyxcgdvo7ugoyxyywe.streamlit.app/)to aid clinical decision-making and treatment planning.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Esophageal cancer (EC) represents the seventh most prevalent malignancy globally (Wang et al. 2022). Esophageal cancer accounts for over 500,000 annual mortalities, constituting 5.3% of all cancer-induced deaths globally. The distribution of this malignancy reveals significant geographical disparities (Bray et al. 2018). The early clinical manifestations of esophageal cancer are insidious and difficult to detect; advanced esophageal cancer presents as progressive dysphagia (Ilson and Hillegersberg 2018). The high mortality rate of esophageal cancer is due to the fact that the majority of patients with esophageal cancer are diagnosed at a late stage, which often leads to the delay in treatment (Uhlenhopp et al. 2020). The pathological types of esophageal cancer are mainly divided into esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) (Njei et al. 2016).
Some studies indicated that esophageal squamous cell carcinoma cases were located in the middle and lower third of the esophagus, while most esophageal adenocarcinomas were located in the lower third (Gasmelseed et al. 2015). Multiple studies have shown that heavy alcohol, smoking, obesity, hot drinks, malnutrition and infections (such as human papillomavirus) can lead to esophageal cancer (Chen et al. 2015; Huang and Yu 2018; Hoyo et al. 2012; Corley et al. 2008; Lindkvist et al. 2014 Feb; Steffen et al. 2009). Metastatic sites of esophageal cancer encompass the liver, brain, lungs, bones, and others. However, lung metastasis in esophageal cancer engenders a substantial impact on patient prognosis. Not only does it signal advanced-stage disease, but it also portends a poor prognosis, resulting in metabolic disorders due to liver dysfunction, circulatory problems. Lung metastasis of esophageal cancer often leads to metabolic disorders, respiratory failure, pain and even multiple organ dysfunction syndrome (MODS) (Luo 2022).
Machine learning (ML) is a branch of data science and an important field of artificial intelligence. Machine learning is mainly based on the development and training of algorithms through computers, ML can learn from data and perform predictions without specific programming before (Choy et al. 2018). The main advantage of ML is that it can analyze and use a large amount of data, and can use more optimized algorithms to build more accurate models. Compared with traditional statistical analysis, ML is much more effective (Gillies et al. 2016). At present, ML technology has been widely used in different fields, from self-driving cars, board games, and various event decisions (Silver et al. 2018).
In clinical medicine, biomedicine and other fields, ML can well deal with various types of big data in scientific research, and help clinicians better understand and predict the disease studied. Therefore, ML has been used in clinical diagnosis, precision treatment and health monitoring (Aarestrup et al. 2020; Zhuang et al. 2020; Shilo et al. 2020).
Patients with esophageal cancer often have different clinical manifestations, pathological grading and metastasis site (Gong et al. 2021). After receiving different treatments, the prognosis results also have significant differences. Unfortunately, there are limited studies on lung metastasis of advanced esophageal cancer, which poses new challenges to clinicians' clinical decision-making. Therefore, the purpose of this study is to use ML to build and verify a new machine learning model to predict lung metastasis in patients with esophageal cancer.
Materials and methods
Study population
The SEER*stat 8.4.1 software was utilized in this study to retrieve clinical data of patients diagnosed with esophageal cancer from the SEER database. The study enrolled patients who were diagnosed with esophageal cancer (including squamous cell carcinoma and adenocarcinoma) in the SEER database from 2010 to 2020.
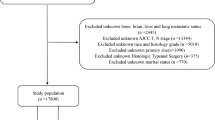
Exclusion criteria were detailed as follows: (1) Excluded unknown race and grade; (2) Excluded unknown primary site; (3) Excluded unknown AJCC T, N stage; (4) Excluded unknown bone, brian, liver and lung metastatic status. The total number of esophageal cancer cases obtained was 20,249, which were subsequently divided into a train set and a test set in a ratio of 7:3. Please refer to Fig. 1 for the complete screening process.
Data selection
In this study, 12 variables related to the clinicopathological and demographic characteristics of patients were selected for analysis. Demographic variables included age, sex and race. Clinicopathological variables included primary site, tumor histology, tumor grade, T stage, N stage, brain metastasis, bone metastasis, lung metastasis, liver metastasis. According to the ICD-O-3 codes, histological types of esophageal cancere divided into 2 categories, including adenocarcinoma (8140–8573), squamous cell carcinoma (8050–8082). All esophageal cancer patients were staged according the AJCC 8th edition guidelines and SEER staging information. Reveal the disparities among various factors in the training and validation sets through chi-square analysis. The age is typically represented by its mean value.
Data pre-processing and feature engineering
All statistical analyses were performed using Python 3.8 and SPSS 23. The logistic regression analysis was conducted on the data collected from the SEER database using SPSS 23 software, aiming to identify variables suitable for machine learning models. The significant variables (P < 0.05) in patients with pulmonary metastasis were identified using univariate logistic regression analysis. Subsequently, these variables were included in multivariate logistic regression analysis, and the variables with P < 0.05 in the multivariate logistic regression analysis underwent further analysis using a machine learning model. The correlation between the selected features was examined using a method of correlation analysis. Since this data set is an unbalanced data set, the over-sampling method were adopted for data processing (Solihah et al. 2020). The key of this method is to oversampling the data samples of small classes to increase the number of data samples of small classes to improve the accuracy of the model. Meanwhile, to compare the importance of each feature, we extract the feature importance of each variable in the machine learning model according to the Permutation Importance principle (Tian et al. 2021; Liu et al. 2021a).
Model establishment and evaluation
The data from the SEER database was randomly assigned in a 7:3 ratio to the train set and the internal test set. Six commonly used classifier algorithms were chosen to this study, including three ensemble algorithms (Random Forest (RF), Gradient Boosting ine (GBM), eXtreme gradient boosting (XGB)) and three simple classification algorithms (Logistic Regression (LR), Decision tree (DT), Naive Bayes classifiers (NBC)). The ML model was trained using Python software. In the train set, all SEER data were split into 10 parts and cross-validated 10 times (Buch et al. 2018). The built model directly imports data for validation in the case of internal test sets. The area under the receiver operating characteristic curve (AUC), sensitivity, specificity, accuracy and F-score were evaluated indicators of ML algorithms. The probability density plot and clinical utility curve (CUC) was utilized to examine clinical applicability. Furthermore, based on the best-performing model, we built a web-based online calculator.
Results
Clinical characteristics of patients
A total of 20,249 patients diagnosed with esophageal cancer were retrieved from the SEER database and subsequently divided into a training set (n = 14,174) and a test set (n = 6,075) in a ratio of 7:3. Twelve factors, including age, gender, race, primary site, histology, tumor grade, T stage, N stage, brain metastasis status as well as bone metastasis status along with lung and liver metastases were collected. The mean age of patients in the training set was 67.2 years (67.2 ± 11.0), whereas the mean age of patients in the validation set was 67.0 years (67.0 ± 11.1). The majority of patients diagnosed with esophageal cancer were male; white individuals constituted the largest proportion; the most prevalent tumor location was observed in the lower third of the esophagus while adenocarcinoma represented the predominant histological type. No significant variation was found in terms of tumor grade distribution among patients. Following chi-square analysis between each factor within both sets (training and validation), no statistically significant differences were identified (p > 0.05). The distribution pattern for each factor exhibited similarity between these two groups which indicates that both the training set and validation set are suitable for further predictive analysis (Table 1).
Univariable and multivariable logistic regression analysis
In our study, 8 risk factors associated with lung metastasis were identified through univariate and multivariate logistic regression analysis, including primary site, tumor histology, tumor grade, T stage, N stage, brain metastasis, bone metastasis and liver metastasis (P < 0.05; Table 2). Based on these 8 independent prognostic factors we developed six different models using machine learning algorithms in this study.
Correlation analysis and Importance of features on prediction
In order to assess the level of correlation among various factors, it is customary to employ correlation analysis. For this study, we utilize Spearman correlation analysis to evaluate the interrelationships and characteristics among the aforementioned 11 factors. A corresponding heat map is generated, as depicted in Figure A The findings from Fig. 2A indicate a lack of significant correlation among the studied 11 characteristics. Given the absence of substantial correlations between these factors, statistical interference is minimized, enabling us to effectively incorporate these factors into constructing a predictive model. he significance of various factors from the six machine learning algorithms is illustrated in Fig. 2B In the RF model, liver metastasis, T stage, and N stage emerge as the top three most influential factors. Similarly, for the XGB model, liver metastasis, bone metastasis, and N stage are deemed to be highly important. The GBM model highlights liver metastasis, T stage, and bone metastasis as its key predictors. In contrast, for the LR model, liver metastasis holds utmost importance along with bone metastasis and brain metastasis. For the DT model, liver metastasis again takes precedence followed by T stage and N stage as significant factors. Lastly, in the NB model, T stage, N Stage, and Grade are considered to be pivotal factors. The analysis reveals that except for the NB model, liver metastasis consistently recognized as the most critical factor. T stage, N stage, bone metastasis, and liver metastasis have been identified as important determinants.
Model performance
The performance of the six predictive models is described in Fig. 3A, B and C Table 3. Internal ten-fold cross-validation (Fig. 3A) showed that GBM model performed best among the six models with an average AUC of 0.893, followed by the LR model (AUC = 0.828). Internal test validation was shown in Table 3 and Fig. 3B. Interestingly, the GBM model also achieves the best AUC score (0.803) in the internal test validation and the score of accuracy, sensitivity (recall rate) and specificity were 0.849, 0.604 and 0.867, respectively. The confusion matrix (Fig. 3C) of the GBM model in the training set and the test set indicated its high accuracy. The probability density plot (Fig. 3D) depicting predictive distribution showed that the AUC was highest when the predictive score was 0.53. The CUC plot (Fig. 3E) also showed good clinical applicability.
A Ten-fold cross-validation results of different machine learning models. B The roc curves of different machine learning models in internal test set. C The confusion matrix of the GBM model in the (A) train set and the (B) internal test set. TP true positive; TN true negative; FP false positive; FN false negative. D Probability density plot of gradient boosting machine model. E The clinical impact curve of gradient boosting machine model
A: Ten-fold cross-validation results of different machine learning models. B: The roc curves of different machine learning models in internal test set. C: The confusion matrix of the GBM model in the (A) train set and the (B) internal test set. TP, true positive; TN, true negative; FP, false positive; FN, false negative. D: Probability density plot of gradient boosting machine model. E: The clinical impact curve of gradient boosting machine model.
Web predictor
The objective of this study was to develop a web predictor utilizing the GBM model, which demonstrated superior predictive performance for lung metastasis in patients with esophageal cancer. The primary aim of this web predictor is to provide healthcare professionals with a valuable tool for making more precise clinical decisions. By inputting the relevant variables associated with hepatic metastasis into the web predictor, doctors can conveniently calculate the probability of lung metastasis in patients with esophageal cancer. Please refer to Supplementary Information Figure for further details. For easy accessibility, please visit the following link to access the web predictor: (https://lvgrkyxcgdvo7ugoyxyywe.streamlit.app/).
Discussion
Esophageal cancer is considered one of the most aggressive malignancies among all gastrointestinal tumors, and it ranks as the most prevalent form of cancer worldwide (Global Burden of Disease Cancer Collaboration 1990). Despite advancements in multidisciplinary treatment approaches for esophageal cancer in the United States, the 5-year relative survival rate remains low at only 20% (Siegel et al. 2022; Watanabe et al. 2020). Notably, there is currently no universally recognized and effective comprehensive systemic therapy available for esophageal cancer with distant metastasis. The anticipated incidence of distant metastasis in clinically detected cases ranges from 27.3 to 66.7% (Lou et al. 2013; Ichida et al. 2013). It is well-known that lung represents one of the primary sites for esophageal cancer metastasis (Kudou et al. 2022).
Both the esophagus and lungs are organs located in the thoracic cavity, playing a crucial role in maintaining normal physiological function, nutrient absorption, and metabolism of the human body. Once lung metastasis occurs in patients with esophageal cancer, it not only indicates disease progression to an advanced stage but also leads to gradual loss of pulmonary function, resulting in respiratory distress, decreased oxygenation index, and potential complications such as malignant pleural effusion and cachexia (Al-Sawaf et al. 2023). The effective treatment and comprehensive management of metastatic esophageal cancer necessitate a multidisciplinary approach encompassing various treatment modalities and strategies. This remains a significant challenge within the medical community. Therefore, identifying high-risk factors for esophageal cancer and accurately predicting the likelihood of lung metastasis based on clinical and pathological characteristics hold immense importance for clinical decision-making.
Existing studies on lung metastasis of esophageal cancer patients remain limited. To the best of our knowledge, there is a lack of research on the development of a prediction model for pulmonary metastasis in esophageal cancer. In order to address this research gap, three key issues need to be addressed. Firstly, it is crucial to identify high-risk prognostic factors associated with esophageal cancer in order to gain insights into its pathogenesis, etiological factors and improve prognosis. Secondly, further investigation into the interrelationships among these independent prognostic factors is also essential. Lastly, leveraging big data and machine learning techniques can enhance the accuracy and effectiveness of predictive modeling in this field. Our study successfully addresses these challenges by utilizing a comprehensive dataset from the SEER database to identify independent prognostic factors related to esophageal cancer and exploring their correlations. Furthermore, we have developed a user-friendly website that allows direct access to our prediction model.
Some studies have indicated that smoking and alcohol consumption are the predominant risk factors for male esophageal cancer (Li et al. 2021). Furthermore, research has demonstrated that in patients with esophageal cancer, the extent of tissue differentiation, pathological stage, vascular invasion, and nerve invasion are widely acknowledged as crucial prognostic factors (Petrelli et al. 2021; Gao et al. 2016; Shahbaz Sarwar et al. 2010; Yang et al. 2020; Gupta et al. 2018). The aforementioned literature comprehensively describes and statistically analyzes the independent prognostic factors of esophageal cancer. However, these studies often lack the support of extensive data, which may compromise their reliability. Furthermore, these studies fail to predict the occurrence of metastasis in esophageal cancer. In contrast, this study utilizes big data analysis from the SEER database to identify independent high-risk factors associated with lung metastasis through logistic regression analysis. This approach effectively mitigates statistical errors caused by small sample sizes. Specifically, this study includes 11 common clinical factors related to lung metastasis: age, sex, race, primary site, tumor histology, tumor grade, brain metastasis, bone metastasis, liver metastasis, T stage, and N stage. In order to ascertain the independence among different features,a correlation heat map was generated using Spearman correlation analysis (Fig. 2). The results depicted in Fig. 2 indicate that there is no significant correlation observed among the 11 features. Subsequently, the univariate and multivariate logistic regression analyses were employed to identify eight independent high-risk factors associated with lung metastasis. These factors include liver metastasis,T stage,N stage,bone metastasis,tumor grade,primary site,tumor histology,and bone metastases.
Undoubtedly, establishing a predictive model for distant organ metastasis in advanced esophageal cancer is of paramount importance in order to investigate independent high-risk factors. Currently, there is an insufficient amount of research on risk factors in patients with esophageal cancer who develop distant organ metastasis (Ai et al. 2019).
For instance, Tang et al. previously constructed a nomogram to predict survival in patients with metastatic esophageal cancer; however, that study encompassed all anatomical sites of metastasis and did not specifically explore models for predicting the risk of distant metastasis (Tang et al. 2019a). Similarly, Cheng et al. developed a customized model for predicting the risk and survival outcomes in patients with esophageal cancer presenting brain metastases (Cheng et al. 2021). Furthermore, Guo et al. provided comprehensive characteristics of patients with liver metastases and investigated risk factors as well as prognostic factors; nevertheless, they did not develop any predictive tools (Guo et al. 2021).
The comprehensive investigation of patients with esophageal cancer and lung metastasis holds significant clinical importance, given the fact that lung metastasis serves as the most prevalent distant dissemination site for this disease.
Previous studies have utilized the nomogram method to develop a prognostic model for patients with esophageal cancer at different stages; however, these studies did not establish a prediction model specifically for late-stage metastatic esophageal cancer (Domper Arnal et al. 2015). Earlier literature has constructed a nomogram based on traditional logistic regression models to predict esophageal cancer metastasis. Nevertheless, the accuracy of this nomogram is limited due to its inability to effectively handle big data. In contrast, current cutting-edge medical research is centered around precision medicine, where the use of nomograms has posed challenges in achieving significant breakthroughs (Deo 2015; Goecks et al. 2020). Furthermore, conventional research methods fail to explore interactions among various independent high-risk factors (Liu et al. 2021b; Tang et al. 2019b). Conversely, our study not only comprehensively captures complex associations between different independent high-risk factors but also employs advanced machine learning statistical techniques to construct an improved prediction model. The model can be considered effective when the AUC value exceeds 0.7 in general (Yu et al. 2021). Our GBM model achieved an AUC value of 0.803, providing strong evidence for the high reliability of our proposed model.
After employing machine learning techniques, we constructed six prediction models and conducted internal tenfold cross-validation to determine the optimal model among them, which turned out to be the GBM model. Utilizing these findings, we successfully developed a publicly accessible online calculator based on the GBM model (https://lvgrkyxcgdvo7ugoyxyywe.streamlit.app/). The meticulously crafted model accurately forecasts the risk of lung metastasis in patients by considering diverse clinical indicators. Clinicians can conveniently access this tool via the provided website, input patient information, and promptly obtain corresponding probabilities of lung metastasis. Consequently, this resource significantly aids clinicians in making informed clinical decisions.
Our study possesses several advantages. Firstly, we have developed a statistical model based on machine learning to accurately predict the probability of pulmonary metastasis in patients with esophageal cancer. To our knowledge, our research team is the first to utilize machine learning for constructing such a prediction model. This model exhibits greater reliability compared to traditional nomogram prediction models. Additionally, this work contributes to expanding our understanding of artificial intelligence and precision medicine. Secondly, our study delves deeper into exploring the relationship between various independent high-risk factors among patients with esophageal cancer, thereby providing new avenues for future clinical research. In other words, future studies should not solely focus on examining the final outcomes of patients but also investigate the correlations between different independent high-risk factors. This approach will enable us to better comprehend these relationships and subsequently eliminate factors that may hinder patients' perioperative lifestyle or treatment methods.
However, it is important to acknowledge certain limitations within this study. Firstly, being a single-center study with a limited number of included patients, we employed internal validation for model verification purposes. Therefore, in subsequent investigations, we plan to incorporate multi-center data for training and external validation in order to obtain an even more reliable prediction model. Secondly and regrettably, Neoadjuvant therapy, surgical methods, circulating tumor DNA and other factors that may affect the long-term prognosis of patients with esophageal cancer were not included in this study. In the future, with the continuous improvement of the database, we will include more parameters related to esophageal cancer into the prediction model to improve its accuracy.
Conclusion
In conclusion, based on eight commonly observed clinicopathological features in clinical practice, this study has successfully developed a machine learning model to accurately predict the occurrence of pulmonary metastasis in patients with esophageal cancer. Among these models, the GBM model demonstrated superior performance. By utilizing the GBM model, clinicians can obtain valuable information regarding the probability of pulmonary metastasis in patients with esophageal cancer, thereby facilitating the development of more precise treatment strategies.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Aarestrup FM, Albeyatti A, Armitage WJ, Auffray C, Augello L, Balling R et al (2020) Towards a European health research and innovation cloud (HRIC). Genome Med 12:18
Ai D, Chen Y, Liu Q, Deng J, Zhao K (2019) The effect of tumor locations of esophageal cancer on the metastasis to liver or lung. J Thorac Dis 11:4205–4210
Al-Sawaf O, Weiss J, Skrzypski M et al (2023) Body composition and lung cancer-associated cachexia in TRACERx. Nat Med 29(4):846–858
Bray F, Ferlay J, Soerjomataram I et al (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Buch VH, Ahmed I, Maruthappu M (2018) Artificial intelligence in medicine: Current trends and future possibilities. Br J Gen Pract 68(668):143–144
Chen W, Zheng R, Zeng H et al (2015) Annual report on status of cancer in China, 2011. Chin J Cancer Res 27:2–12
Cheng S, Yang L, Dai X, Wang J, Han X (2021) The risk and prognostic factors for brain metastases in esophageal cancer patients: an analysis of the SEER database. BMC Cancer 21:1057
Choy G, Khalilzadeh O, Michalski M et al (2018) Current applications and future impact of machine learning in radiology. Radiology 288(2):318–328
Corley DA, Kubo A, Zhao W (2008) Abdominal obesity and the risk of esophageal and gastric cardia carcinomas. Cancer Epidemiol Biomarkers Prev 17(2):352–358
Deo RC (2015) Machine learning in medicine. Circulation 132:1920–1930
DomperArnal MJ, Ferrández Arenas Á, Lanas AÁ (2015) Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol 21(26):7933–7943
Gao A, Wang L, Li J et al (2016) Prognostic value of perineural invasion in esophageal and esophagogastric junction carcinoma: a metaanalysis. Dis Markers 2016:7340180
Gasmelseed N, Abudris D, Elhaj A et al (2015) Patterns of esophageal cancer in the National Cancer Institute at the University of Gezira, in Gezira State, Sudan, in 1999–2012. Asian Pac J Cancer Prev 16(15):6481–6490
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are DATA. Radiology 278(2):563–577
Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Abate D et al (2019) Global, regional, and national cancer incidence. JAMA Oncol 5(12):1749–1768
Goecks J, Jalili V, Heiser LM, Gray JW (2020) How machine learning will transform biomedicine. Cell 181:92–101
Gong X, Zheng B, Xu G et al (2021) Application of machine learning approaches to predict the 5-year survival status of patients with esophageal cancer. J Thorac Dis 13(11):6240–6251
Guo J, Zhang S, Li H, Hassan MOO, Lu T, Zhao J et al (2021) Lung metastases in newly diagnosed esophageal cancer: a population-based study. Front Oncol 11:603953
Gupta V, Coburn N, Kidane B et al (2018) Survival prediction tools for esophageal and gastroesophageal junction cancer: a systematic review. J Thorac Cardiovasc Surg 156(2):847–856
Hoyo C, Cook MB, Kamangar F et al (2012) Body mass index in relation to oesophageal and oesophagogastric junction adenocarcinomas: a pooled analysis from the International BEACON Consortium. Int J Epidemiol 41(6):1706–1718
Huang FL, Yu SJ (2018) Esophageal cancer: risk factors, genetic association, and treatment. Asian J Surg 41:210–215
Ichida H, Imamura H, Yoshimoto J et al (2013) Pattern of postoperative recurrence and hepatic and/or pulmonary resection for liver and/or lung metastases from esophageal carcinoma. World J Surg 37(2):398–407
Ilson DH, van Hillegersberg R (2018) Management of patients with adenocarcinoma or squamous cancer of the esophagus. Gastroenterology 154(2):437–451
Kudou K, Saeki H, Nakashima Y et al (2022) Clinical outcomes of surgical resection for recurrent lesion after curative esophagectomy for esophageal squamous cell carcinoma: a nationwide, large-scale retrospective study. Esophagus 19(1):57–68
Li S, Chen H, Man J et al (2021) Changing trends in the disease burden of esophageal cancer in China from 1990 to 2017 and its predicted level in 25 years. Cancer Med 10(5):1889–1899
Lindkvist B, Johansen D, Stocks T et al (2014) Metabolic risk factors for esophageal squamous cell carcinoma and adenocarcinoma: a prospective study of 580,000 subjects within the Me-Can project. BMC Cancer 18(14):103
Liu W-C, Li M-X, Qian W-X, Luo Z-W, Liao W-J, Liu Z-L et al (2021a) Application of machine learning techniques to predict bone metastasis in patients with prostate cancer. Cancer Manag Res 13:8723–8736
Liu X, Guo W, Shi X et al (2021b) Construction and verification of prognostic nomogram for early-onset esophageal cancer. Bosn J Basic Med Sci 21(6):760–772
Lou F, Sima CS, Adusumilli PS et al (2013) Esophageal cancer recurrence patterns and implications for surveillance. J Thorac Oncol 8(12):1558–1562
Luo P, Wei X, Liu C et al (2022) The risk and prognostic factors for liver metastases in esophageal cancer patients: a large-cohort based study. Thorac Cancer 13(21):2960
Njei B, McCarty TR, Birk JW (2016) Trends in esophageal cancer survival in United States adults from 1973 to 2009: a SEER database analysis. J Gastroenterol Hepatol 31(6):1141–1146
Petrelli F, Ghidini A, Cabiddu M et al (2021) Effects of hypertension on cancer survival: a meta-analysis. Eur J Clin Invest 51(6):e13493
ShahbazSarwar CM, Luketich JD, Landreneau RJ et al (2010) Esophageal cancer: an update. Int J Surg 8(6):417–422
Shilo S, Rossman H, Segal E (2020) Axes of a revolution: Challenges and promises of big data in healthcare. Nat Med 26:29–38
Siegel RL, Miller KD, Fuchs HE et al (2022) Cancer statistics, 2022. CA Cancer J Clin 72(1):7–33
Silver D, Hubert T, Schrittwieser J, Antonoglou I, Lai M, Guez A et al (2018) A general reinforcement learning algorithm that masters chess, shogi, and go through self-play. Science 362:1140–1144
Solihah B, Azhari A, Musdholifah A (2020) Enhancement of conformational b-cell epitope prediction using CluSMOTE. PeerJ Comput Sci 6:e275
Steffen A, Schulze MB, Pischon T et al (2009) Anthropometry and esophageal cancer risk in the European prospective investigation into cancer and nutrition. Cancer Epidemiol Biomarkers Prev 18(7):2079–2089
Tang X, Zhou XJ, Li YY, Tian X, Wang Y, Huang M et al (2019a) A novel nomogram and risk classification system predicting the cancerspecific survival of patients with initially diagnosed metastatic esophageal cancer: a SEER-based study. Ann Surg Oncol 26:321–328
Tang X, Zhou X, Li Y et al (2019b) A novel nomogram and risk classification system predicting the cancer-specific survival of patients with initially diagnosed metastatic esophageal cancer: A SEER-based study. Ann Surg Oncol 26(2):321–328
Tian H, Ning Z, Zong Z, Liu J, Hu C, Ying H et al (2021) Application of machine learning algorithms to predict lymph node metastasis in early gastric cancer. Front Med (Lausanne) 8:759013
Uhlenhopp DJ, Then EO, Sunkara T et al (2020) Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin J Gastroenterol 13(6):1010–1021
Wang R, Liu S, Chen B et al (2022) Recent advances in combination of immunotherapy and chemoradiotherapy for locally advanced esophageal squamous cell carcinoma. Cancers (Basel) 14(20):5168
Watanabe M, Otake R, Kozuki R et al (2020) Recent progress in multidisciplinary treatment for patients with esophageal cancer. Surg Today 50(1):12–20
Yang J, Lu Z, Li L et al (2020) Relationship of lymphovascular invasion with lymph node metastasis and prognosis in superficial esophageal carcinoma: Systematic review and meta-analysis. BMC Cancer 20(1):176
Yu J, Hu W, Yao N, Sun M, Li X, Wang L et al (2021) Development and validation of a nomogram to predict overall survival of T1 esophageal squamous cell carcinoma patients with lymph node metastasis. Transl Oncol 14:101127
Zhuang Y, Chen YW, Shae ZY, Shyu CR (2020) Generalizable layered blockchain architecture for health care applications: development, case studies, and evaluation. J Med Internet Res 22:e19029
Acknowledgements
We thank all patients, investigators, and institutions involved in this study, especially the SEER database.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Jun Wan and Yukai Zeng designed the experiments. Jun Wan collected and processed the data. Yukai Zeng wrote and polished article. All of the authors read and approved the final manuscript. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval and consent to participate
Not applicable.
Consent for publication
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Research involving human and animal rights
The authors stated that no human or animal experiments were adopted in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fang, Y., Wan, J. & Zeng, Y. Use machine learning to predict pulmonary metastasis of esophageal cancer: a population-based study. J Cancer Res Clin Oncol 150, 420 (2024). https://doi.org/10.1007/s00432-024-05937-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00432-024-05937-6