Abstract
Background
[18F]Fluoro-deoxyglucose (FDG) positron emission tomography/computed tomography (PET/CT) is the standard imaging procedure in diffuse large B-cell lymphoma (DLBCL). Disease presentation, FDG-PET/CT performance, and outcome may be influenced by germline single nucleotide polymorphisms (SNP) in genes regulating glucose uptake.
Methods
Clinical variables, FDG-PET findings, and outcome were analysed in relation to SNPs in 342 DLBCL patients participating in the ‘Positron Emission Tomography-Guided Therapy of Aggressive Non-Hodgkin Lymphomas’ (PETAL) trial. Genes analysed included SLC2A1 (SNPs rs1385129, referred to as HaeIII; rs710218, HpyCH4V; rs841853, XbaI), VEGFA (rs3025039), HIF1A (rs11549465, P582S; rs11549467, A588T), and APEX1 (rs1130409, D148E). Statistical significance was assumed at p ≤ 0.05.
Results
The SLC2A1 HaeIII and HpyCH4V SNPs were tightly linked and statistically significantly associated with baseline maximum standardized uptake value (SUVmax) and Ann Arbor stage, with slightly lower SUVmax (HaeIII, median 18.9, interquartile range [IQR] 11.5–26.6, versus 21.6, IQR 14.4–29.7; p = 0.019) and more frequent stage IV disease (HaeIII, 44.5% versus 30.8%; p = 0.011) in minor allele carriers. As previously reported for lung cancer, the association was dependent upon the coexistent APEX1 D148E genotype. The HIF1A A588T SNP was associated with total metabolic tumour volume (TMTV) and time-to-progression, with significantly lower TMTV (median 16 cm3, IQR 7–210, versus 146 cm3, IQR 34–510; p = 0.034) and longer time-to-progression in minor allele carriers (log-rank p = 0.094). Time-to-progression was also associated with the SLC2A1 XbaI and APEX1 D148E SNPs, with shorter time-to-progression in homozygous and heterozygous SLC2A1 XbaI (HR 1.456; CI 0.930–2.280; p = 0.099) and homozygous APEX1 D148E minor allele carriers (HR 1.6; CI 1.005–2.545; p = 0.046). In multivariable analyses including SNPs, International Prognostic Index factors, sex, and B symptoms, HIF1A A588T, SLC2A1 XbaI, and APEX1 D148E retained statistical significance for time-to-progression, and SLC2A1 XbaI was also significantly associated with overall survival.
Conclusions
Common SNPs in genes regulating glucose uptake may impact SUVmax, tumour distribution, tumour volume, and outcome in DLBCL. The effects on SUVmax are of low magnitude and appear clinically negligible. The results are consistent with findings in other types of cancer. They need to be confirmed in an independent DLBCL population of sufficient size.
Trial registration
Trial registration: ClinicalTrials.gov NCT00554164; EudraCT 2006-001641-33. Registration date November 5, 2007, https://www.clinicaltrials.gov/ct2/show/NCT00554164
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Diffuse large B-cell lymphoma (DLBCL) is the most frequent cancer of the immune system (Morton et al. 2006). [18F]Fluoro-deoxyglucose (FDG) positron emission tomography/computed tomography (PET/CT) is the standard imaging procedure to define tumour stage and treatment response (Cheson et al. 2014). Because stage and response may impact therapy (Poeschel et al. 2020; Persky et al. 2020), reliable FDG-PET/CT performance is of utmost importance.
The uptake of glucose and FDG is controlled by several genes. One of the most important ones is solute carrier family 2 member 1 (SLC2A1, also known as GLUT1, facilitated glucose transporter type 1) encoding the major channel for glucose entry into the cell (Ancey et al. 2018). The expression of SLC2A1 is stimulated by the transcription factor hypoxia-inducible factor 1α (gene HIF1A), the major regulator of hypoxic responses (Lee et al. 2019). Another HIF1α target gene is VEGFA whose protein product, vascular endothelial growth factor A, induces blood vessel formation, thus increasing glucose supply (Apte et al. 2019). The activity of HIF1α is enhanced by apurinic/apyrimidinic endonuclease 1 (APEX1), a multifunctional protein involved in DNA repair and transcription factor activation (Shah et al. 2017). Single nucleotide polymorphisms (SNP) in these genes have been studied extensively with regard to risk, presentation, and outcome of cancer (Amann et al. 2011; Feng et al. 2017; Heist et al. 2008; Kim et al. 2012; Tanimoto et al. 2003; Knechtel et al. 2010; Qin et al. 2012; Hill et al. 2006; Matakidou et al . 2007; Smith et al. 2008; Cao et al. 2011). In some studies, their impact on FDG-PET performance was also evaluated (Wolf et al. 2004; Grabellus et al. 2010; Kim et al. 2010; Bravatà et al. 2013).
We investigated the impact of SNPs in the named four genes on DLBCL presentation, FDG-PET findings, and outcome in the setting of the ‘Positron Emission Tomography-Guided Therapy of Aggressive Non-Hodgkin Lymphomas’ (PETAL) trial (Dührsen et al. 2018). The trial set out – and failed – to improve treatment by adapting it to the response to the first two cycles of therapy. Because interim PET-driven treatment changes had no impact on outcome, all treatment arms were combined in the present analysis.
Methods
Study design
The PETAL trial (ClinicalTrials.gov NCT00554164; EudraCT 2006-001641-33) was a multicentre study for newly diagnosed aggressive non-Hodgkin lymphomas (Dührsen et al. 2018). Diagnoses were confirmed by reference pathological review including molecular analyses (Richter et al. 2019). The study was approved by the Federal Institute for Drugs and Medical Devices and the ethics committees of the participating sites. All patients gave written informed consent including permission of data use for post hoc scientific analyses.
Patients were treated with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP). Interim PET was performed a median of 20 days after cycle 2. Patients with favourable response received four more cycles of R-CHOP or the same treatment plus two extra doses of rituximab. Patients with unfavourable response were randomly assigned to receive six additional cycles of R-CHOP or six blocks of a more intensive protocol originally developed for Burkitt’s lymphoma (Dührsen et al. 2018).
FDG-PET/CT imaging and evaluation
The imaging conditions have been described previously (Dührsen et al. 2018). The treatment response was determined by dividing the maximum standardized uptake value (SUVmax) of the hottest residual lesion on the interim scan by the SUVmax of the hottest lesion on the baseline scan. Favourable scans were defined by complete disappearance of all non-physiological FDG activity or SUVmax reduction by > 66% (Rekowski et al. 2021). During the trial, the treatment response was assessed by local investigators. A post hoc central review yielded a concordance of 97.7% with the original assessment (Dührsen et al. 2018). Total metabolic tumour volume (TMTV) was determined centrally on archived PET/CT scans, using the 41% SUVmax method (Schmitz et al. 2020). End-of-treatment responses were defined by CT criteria (Cheson et al. 2014; Dührsen et al. 2018).
Single nucleotide polymorphisms
The analysis was restricted to glucose uptake-regulating genes whose SNPs had previously been shown to be associated with FDG-PET findings (Wolf et al. 2004; Grabellus et al. 2010; Kim et al. 2010) or tumour mass (Tanimoto et al. 2003; Knechtel et al. 2010; Qin et al. 2012). Germline SNPs in genomic DNA from peripheral blood mononuclear cells were detected by allele-specific polymerase chain reaction (PCR) using Assay-on-demand TaqMan SNP assays that contained primers and allele-specific VIC- and FAM-labelled probes and genotyping master mix (Thermo Fisher Scientific, Waltham, MA, USA). Cycling conditions were 60 °C for 30 s and an initial denaturation step of 95 °C for 10 min followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s using StepOne Plus real-time PCR instrumentation (Thermo Fisher Scientific). Allele typing was performed using the StepOne Software. In every PCR plate, negative controls as well as internal positive controls were included for homozygous and heterozygous sample types.
HIF1A expression
Expression of the HIF1α protein was studied by immunohistochemistry using the antibody H1alpha67 (Novus Biologicals, Abingdon, United Kingdom; 1:50, pH 9), with normoxic and hypoxic BL-70 cells serving as negative and positive controls, respectively. To induce hypoxia, BL-70 cells were grown in the presence of 100 µM CoCl2 for 20 h at 37 °C in 5% CO2 in air, followed by preparation of cell pellets in formalin-fixed paraffin-embedded blocks.
Statistical analysis
All analyses were exploratory, applying a two-sided alpha of 0.05. Numerical variables and frequencies were compared using the Kruskal–Wallis and chi2 tests, respectively, and time-to-event end-points were analysed using the Kaplan–Meier estimator, the log-rank test, and, when adjusting for covariates, Cox proportional hazards regression. Results were not corrected for multiple testing. All analyses were carried out using IBM SPSS Statistics, version 26.0, Armonk, NY, USA.
Results
Patient characteristics
Of 862 patients treated in the PETAL trial, 609 had DLBCL (Dührsen et al. 2018). Peripheral blood for genotyping was collected from 342 patients (Table 1). Favourable and unfavourable interim PET responses were recorded in 308 (90.1%) and 34 patients (9.9%), respectively.
SUVmax and TMTV were determined in 341 and 298 patients, respectively. Median baseline SUVmax was 20.5 (interquartile range [IQR], 13.3–28.8), median SUVmax reduction at interim scanning was 83.1% (IQR, 73.1–88.5), and median baseline TMTV was 140 cm3 (IQR, 33–494). TMTV was below or above the previously defined prognostic threshold of 328 cm3 (Schmitz et al. 2020) in 196 (65.8%) and 102 patients (34.2%), respectively.
Reference pathological review was performed in 337 patients (98.5%). Cell-of-origin-based subtyping using the Hans classifier (Hans et al. 2004) was available for 162 lymphomas, showing germinal centre B-cell (GCB) and non-GCB lymphomas in 80 (49.4%) and 82 cases (50.6%), respectively. The double-hit status (simultaneous translocations of MYC and BCL2 or BCL6) was determined in 146 lymphomas, with a positive result in 11 cases (7.5%).
SNP characteristics
Genotyping included seven commonly studied SNPs in SLC2A1 (rs1385129, subsequently referred to as HaeIII; rs710218, HpyCH4V; rs841853, XbaI), VEGFA (rs3025039), HIF1A (rs11549465, P582S; rs11549467, A588T), and APEX1 (rs1130409, D148E). Genotypes and allele frequencies are displayed in the Supplementary Information, Supplementary Table 1. Deviations from the Hardy–Weinberg equilibrium were not observed. The SLC2A1 HaeIII and HpyCH4V SNPs were tightly linked, with 338 of 342 patients (98.8%) showing identical allelic distributions.
SNPs and baseline clinical features
Sex, age, performance status, extranodal disease, serum lactate dehydrogenase, B symptoms (as dichotomized in Table 1), histological bone marrow involvement, International Prognostic Index, cell-of-origin and double-hit status failed to show an association with any of the SNPs tested.
SNPs and FDG-PET/CT findings
The SLC2A1 HaeIII and HpyCH4V genotypes were statistically significantly associated with baseline SUVmax, with lower values in minor allele carriers than in homozygous major allele carriers (HaeIII, median 18.9 versus 21.6, p = 0.019; HpyCH4V, 19.0 versus 21.4, p = 0.030; Table 2). The association of the SLC2A1 HpyCH4V genotype with SUVmax has previously been shown to be dependent upon the APEX1 D148E genotype (Kim et al. 2010). In line with this observation, the association of SLC2A1 HaeIII and HpyCH4V genotypes with SUVmax was restricted to homozygous APEX1 major allele carriers (Supplementary Table 2). Irrespective of the APEX1 genotype, none of the other SNPs including SLC2A1 XbaI showed an association with baseline SUVmax (Supplementary Tables 2 and 3).
The SLC2A1 HaeIII and HpyCH4V genotypes were also significantly associated with Ann Arbor stage (chi2 test, p = 0.006 and p = 0.005, respectively), driven by a higher number of stage IV patients among minor allele carriers than among homozygous major allele carriers (HaeIII, 44.5% versus 30.8%, p = 0.011; HpyCH4V, 44.7% versus 30.5%, p = 0.008; Table 3). The association was restricted to homozygous or heterozygous APEX1 major allele carriers (Supplementary Table 4). None of the other SNPs showed an association with Ann Arbor stage (Supplementary Tables 4 and 5).
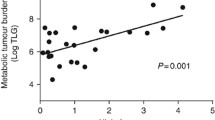
Baseline TMTV was associated with the HIF1A A588T SNP, with significantly lower volumes in minor allele carriers. Because data on the SNP’s functional consequences are scant, we studied HIF1α expression by immunohistochemistry in tumour samples from major and minor allele carriers. One of 4 minor allele carriers and 3 of 16 homozygous major allele controls matched for TMTV (median, 17 cm3 vs. 18 cm3; range, 6–89 vs. 4–147), cell-of-origin, and double-hit status showed weak staining for HIF1α (p = 1, Fisher’s exact test; Fig. 1). A statistically significant association was also found between TMTV and the HIF1A P582S genotype, but this finding was not further pursued, as it was based on only two patients (Table 4). None of the other SNPs was correlated with TMTV (Supplementary Table 6).
HIF1α protein expression by immunohistochemistry using the H1alpha67 antibody. A No expression in normoxic, and B strong expression (brown) in hypoxic BL-70 controls. C No expression in the majority of tumour cells in a heterozygous HIF1A A588T minor allele carrier, with (D), some positive cells in an area close to necrosis. E < 1% scattered positive cells, and F < 10% positive cells in homozygous HIF1A A588T major allele carriers. The bar is equal to 20 µm
SUVmax reduction at interim scanning and interim PET response failed to show an association with SNPs (Supplementary Tables 7 and 8).
SNPs and outcome
Outcome measures included end-of-treatment remission status, time-to-progression, and overall survival. There was no correlation between SNPs and remission status (Table 1). With a median follow-up of 52 months, a trend (p < 0.1) for reduced time-to-progression was observed in homozygous or heterozygous minor allele carriers of the SLC2A1 XbaI SNP and in homozygous minor allele carriers of the APEX1 D148E SNP (Fig. 2). Time-to-progression was prolonged in minor allele carriers of the HIF1A A588T SNP. Overall survival showed a weak correlation with SLC2A1 XbaI and HIF1A A588T, which was more pronounced in homozygous SLC2A1 XbaI minor allele carriers (p = 0.082; data not shown).
To investigate the SNPs’ impact in relation to other variables, they were entered into multivariable Cox regression analyses with stepwise backward elimination of factors failing statistical significance. The first step included SNPs, sex, age, performance status, Ann Arbor stage, extranodal disease, serum lactate dehydrogenase, and B symptoms as dichotomized in Table 1. The final models were dominated by clinical variables, but SLC2A1 XbaI, APEX1 D148E, and HIF1A A588T were retained in the time-to-progression model and SLC2A1 XbaI was retained in the overall survival model (Table 5). Similar results were obtained when the model included the International Prognostic Index risk groups (Table 1) instead of the five factors defining the index (data not shown).
When TMTV dichotomized at 328 cm3 (Schmitz et al. 2020) was added to a model restricted to patients with available TMTV data, HIF1A A588T lost its significance for time-to-progression (p = 0.118), while SLC2A1 XbaI and APEX1 D148E were retained at borderline significance (p = 0.054). SLC2A1 XbaI was also retained in the overall survival model (p = 0.014; Supplementary Table 9).
Discussion
Key findings of our study include the association of the SLC2A1 HaeIII and HpyCH4V SNPs with baseline SUVmax and Ann Arbor stage, the association of the HIF1A A588T SNP with TMTV, and the association of the HIF1A A588T, SLC2A1 XbaI and APEX1 D148E SNPs with long-term outcome.
The SLC2A1 HaeIII and HpyCH4V SNPs were tightly linked, yielding almost identical results. The HpyCH4V SNP is located in the SLC2A1 promotor region in proximity to a putative hypoxic response element which may affect HIF1α binding and SLC2A1 expression (Feng et al. 2017). The minor allele was found to be associated with increased expression in colorectal cancer (Feng et al. 2017) and decreased expression in hepatocellular carcinoma (Amann et al. 2011). In breast cancer (Grabellus et al. 2010; Bravatà et al. 2013) and non-small cell lung cancer (Kim et al. 2010), baseline SUVmax was not affected by the SNPs, but when the analysis was restricted to lung cancer patients homozygous for the APEX1 D148E major allele, homozygosity for the SLC2A1 HpyCH4V minor allele was significantly associated with increased SUVmax (Kim et al. 2010). The APEX1 genotype dependency of the SLC2A1 SNPs’ association with SUVmax was confirmed in our study. However, the variant alleles were associated with decreased rather than increased SUVmax, both in homozygous and heterozygous patients. As pointed out above, the direction in which the variant allele shifts SLC2A1 expression varies from cancer to cancer (Amann et al. 2011; Feng et al. 2017). This may also hold true for the SLC2A1/APEX1 interaction whose nature has yet to be elucidated. In contrast to HaeIII and HpyCH4V, the SLC2A1 XbaI SNP was not correlated with SUVmax. This finding is at variance with a breast cancer study, where homozygosity for the major allele was associated with increased SUVmax (Grabellus et al. 2010).
The SLC2A1 HaeIII and HpyCH4V SNPs were also associated with Ann Arbor stage, a well-established measure of lymphoma distribution (Cheson et al. 2014). Stage IV was overrepresented in HaeIII and HpyCH4V minor allele carriers, provided they also carried the APEX1 major allele. This unexpected association would find an explanation if disseminated disease were more readily detectable in HaeIII or HpyCH4V minor allele carriers. Two observations argue against this assumption. First, SUVmax was lower in minor allele carriers than in homozygous major allele carriers, making improved detectability of low intensity lesions unlikely. Second, in contrast to the association with SUVmax, the association with stage IV disease was seen in both homozygous and heterozygous APEX1 major allele carriers, suggesting distinct biological principles.
Minor allele carriers of the HIF1A A588T SNP had significantly reduced TMTV, a measure of tumour mass. Irrespective of genotype, HIF1α protein expression was undetectable or weak by immunohistochemistry. Since the amino acid substitution has been reported to increase the protein’s transactivation capacity, enhanced function does not require increased expression (Tanimoto et al. 2003). HIF1α-mediated effects on tumour mass have been demonstrated previously (Schwab et al. 2012). Similar to our observation, renal cell carcinoma patients with variant HIF1A alleles including A588T were shown to present in low tumour stages (Qin et al. 2012). By contrast, in head-and-neck and colorectal cancer, tumour size was found to be increased (Tanimoto et al. 2003; Knechtel et al. 2010). The direction of the effect of a variant HIF1A allele on tumour mass appears to be cancer type-specific.
To assess the SNPs’ impact on long-term outcome, we chose two non-overlapping endpoints. Time-to-progression best captures disease-related features, such as disease course and response to therapy. Overall survival is dominated by patient features, such as age, comorbidities, and disease and treatment tolerance (Schmitz et al. 2020). The HIF1A A588T minor allele had a pronounced effect on both endpoints, but this observation was based on only ten patients. Importantly, the SNP’s impact on time-to-progression lost its significance, when TMTV was included in the prognostic model, suggesting that the minor allele’s beneficial effect on outcome was mediated by low tumour mass. In line with our observation, survival was also prolonged in renal cell carcinoma patients carrying a variant allele (Qin et al. 2012). Irrespective of SNPs, detectable HIF1α expression has been reported to portend a favourable prognosis in DLBCL (Evens et al. 2010). In the small set studied here, none of 4 patients with and 2 of 16 patients without detectable HIF1α expression progressed (time-to-progression, log-rank p = 0.517; data not shown).
Time-to-progression was also affected by SLC2A1 XbaI and APEX1 D148E. With regard to the former, a detrimental effect of the minor allele was demonstrable in both homozygous and heterozygous carriers, while with the latter, this was restricted to homozygous patients. In contrast to all other SNPs, XbaI not only influenced time-to-progression, but also overall survival. To our knowledge, the biological consequences of the XbaI SNP are unknown, and SNP-related outcome data are not available for other types of cancer. As for APEX1 D148E, the amino acid substitution has been associated with reduced DNA repair and increased genotoxic stress (Lirussi et al. 2016). This is associated with a predisposition for solid tumours (Smith et al. 2008; Cao et al. 2011), but not non-Hodgkin lymphoma (Hill et al. 2006). Functional data on HIF1α regulation by the variant APEX1 protein is lacking. In lung cancer patients homozygous for the APEX1 D148E minor allele, overall survival was reported to be prolonged rather than reduced, again demonstrating differences between cancer types (Matakidou et al 2007).
The VEGFA SNP failed to show a correlation with any of the variables tested. The minor allele is associated with reduced VEGF expression (Wolf et al. 2004), which may explain why its effect on some types of cancer is favourable (Heist et al. 2008; Wolf et al. 2004). Similar to our findings, no impact on outcome was observed in a previous DLBCL study (Kim et al. 2012).
Strengths of our study include its large sample size, prospective nature, and rigorously controlled conditions of PET performance and treatment delivery. Its major limitation is the multitude of SNPs and variables tested. Most of the associations would have failed to reach statistical significance, had they been corrected for multiple testing. Thus, our results are hypothesis-raising rather than definitive. Most of them, however, are consistent with previous findings. SNPs in SLC2A1 influence SUVmax. Though statistically significant, the extent of SNP-related variability appears clinically negligible in DLBCL, not exceeding the expected test–retest variation for SUV measurements (Kurland et al. 2019). SNPs in HIF1A are related to tumour mass, a major determinant of disease outcome (Schmitz et al. 2020). APEX1 D148E is also related to outcome, possibly mediated by disturbed DNA repair. Finally, the direction of the SNP effects on the investigated variables appears to be cancer type-specific. The reason for this observation remains to be elucidated.
Conclusions
Common SNPs in genes regulating glucose uptake may impact SUVmax, tumour distribution, tumour volume, and outcome in DLBCL. The results are consistent with findings in other types of cancer. They need to be confirmed in an independent DLBCL population of sufficient size.
Availability of data and materials
The datasets used and analysed during this study are available from the corresponding author on reasonable request.
Abbreviations
- APEX1:
-
Apurinic/apyrimidinic endonuclease 1
- CT:
-
Computed tomography
- DLBCL:
-
Diffuse large B-cell lymphoma
- FDG:
-
[18F]fluoro-deoxyglucose
- GCB:
-
Germinal centre B-cell
- GLUT1:
-
Facilitated glucose transporter type 1
- HIF1A:
-
Hypoxia-inducible factor 1A (gene)
- HIF1α:
-
Hypoxia-inducible factor 1α (protein)
- IQR:
-
Interquartile range
- PCR:
-
Polymerase chain reaction
- PET:
-
Positron emission tomography
- PETAL:
-
Positron Emission Tomography-Guided Therapy of Aggressive Non-Hodgkin Lymphomas
- R-CHOP:
-
Rituximab, cyclophosphamide, doxorubicin, vincristine, prednisone
- SLC2A1:
-
Solute carrier family 2 member 1
- SNP:
-
Single nucleotide polymorphism
- SUV:
-
Standardized uptake value
- SUVmax :
-
Maximum standardized uptake value
- TMTV:
-
Total metabolic tumour volume
- VEGFA:
-
Vascular endothelial growth factor A
References
Morton LM, Wang SS, Devesa SS, Hartge P, Weisenburger DD, Linet MS (2006) Lymphoma incidence patterns by WHO subtype in the United States, 1992–2001. Blood 107:265–276
Cheson BD, Fisher RI, Barrington SF, Cavalli F, Schwartz LH, Zucca E et al (2014) Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J Clin Oncol 32:3059–3068
Poeschel V, Held G, Ziepert M, Witzens-Harig M, Holte H, Thurner L et al (2020) Four versus six cycles of CHOP chemotherapy in combination with six applications of rituximab in patients with aggressive B-cell lymphoma with favourable prognosis (FLYER): a randomised, phase 3, non-inferiority trial. Lancet 394:2271–2281
Persky DO, Li H, Stephens DM, Park SI, Bartlett NL, Swinnen LJ et al (2020) Positron emission tomography-directed therapy for patients with limited-stage diffuse large B-cell lymphoma: results of Intergroup National Clinical Trials Network Study S1001. J Clin Oncol 38:3003–3011
Ancey P-B, Contat C, Meylan W (2018) Glucose transporters in cancer—from tumor cells to the tumor microenvironment. FEBS J 285:2926–2943
Lee JW, Ko J, Ju C, Eltzschig HK (2019) Hypoxia signaling in human diseases and therapeutic targets. Exp Mol Med 51:1–13
Apte RS, Chen DS, Ferrara N (2019) VEGF in signaling and disease: beyond discovery and development. Cell 176:1248–1264
Shah F, Logsdon D, Richard A, Messmann RA, Fehrenbacher JC, Fishel ML, Kelley MR (2017) Exploiting the Ref-1-APE1 node in cancer signaling and other diseases: from bench to clinic. NPJ Precis Oncol 1:19
Amann T, Kirovski G, Bosserhoff AK, Hellerbrand C (2011) Analysis of a promoter polymorphism of the GLUT1 gene in patients with hepatocellular carcinoma. Mol Membr Biol 28:182–186
Feng W, Cui G, Tang C-W, Zhang X-L, Dai C, Xu Y-Q et al (2017) Role of glucose metabolism related gene GLUT1 in the occurrence and prognosis of colorectal cancer. Oncotarget 8:56850–56857
Heist RS, Zhai R, Liu G, Zhou W, Lin X, Su Li, et al (2008) VEGF polymorphisms and survival in early-stage non-small-cell lung cancer. J Clin Oncol 26:856–62.
Kim MK, Suh C, Chi HS, Cho HS, Bae YK, Lee KH et al (2012) VEGFA and VEGFR2 genetic polymorphisms and survival in patients with diffuse large B cell lymphoma. Cancer Sci 103:497–503
Tanimoto K, Yoshiga K, Eguchi H, Kaneyasu M, Ukon K, Kumazaki T et al (2003) Hypoxia-inducible factor-1α polymorphisms associated with enhanced transactivation capacity, implying clinical significance. Carcinogenesis 24:1779–1783
Knechtel G, Szkandera J, Stotz M, Hofmann G, Langsenlehner U, Krippl P et al (2010) Single nucleotide polymorphisms in the hypoxia-inducible factor-1 gene and colorectal cancer risk. Mol Carcinog 49:805–809
Qin C, Cao Q, Ju X, Wang M, Meng X, Zhu J et al (2012) The polymorphisms in the VHL and HIF1A genes are associated with the prognosis but not the development of renal cell carcinoma. Ann Oncol 23:981–989
Hill DA, Wang SS, Cerhan JR, Davis S, Cozen W, Severson RK et al (2006) Risk of non-Hodgkin lymphoma (NHL) in relation to germline variation in DNA repair and related genes. Blood 108:3161–3167
Matakidou A, El Galta R, Webb EL, Rudd MF, Bridle H, GELCAPS Consortium et al (2007) Genetic variation in the DNA repair genes is predictive of outcome in lung cancer. Hum Mol Genet 16:2333–40
Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, Hoang KN et al (2008) Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis 29:2132–2138
Cao Q, Qin C, Meng X, Ju X, Ding Q, Wang M et al (2011) Genetic polymorphisms in APE1 are associated with renal cell carcinoma risk in a Chinese population. Mol Carcinog 50:863–870
Wolf G, Aigner RM, Schaffler G, Langsenlehner U, Renner W, Samonigg H et al (2004) The 936C>T polymorphism of the gene for vascular endothelial growth factor is associated with 18F-fluorodeoxyglucose uptake. Breast Cancer Res Treat 88:205–208
Grabellus F, Sheu S-Y, Bachmann HS, Lehmann N, Otterbach F, Heusner TA et al (2010) The XbaI G>T polymorphism of the glucose transporter 1 gene modulates 18F-FDG uptake and tumor aggressiveness in breast cancer. J Nucl Med 51:1191–1197
Kim S-J, Hwang S-H, Kim IJ, Lee MK, Lee CH, Lee S-Y, Lee EY (2010) The association of 18F-deoxyglucose (FDG) uptake of PET with polymorphisms in the glucose transporter gene (SLC2A1) and hypoxia-related genes (HIF1A, VEGFA, APEX1) in non-small cell lung cancer. SLC2A1 polymorphisms and FDG-PET in NSCLC patients. J Exp Clin Cancer Res 29:69
Bravatà V, Stefano A, Cammarata FP, Minafra L, Russo G, Nicolosi S et al (2013) Genotyping analysis and 18FDG uptake in breast cancer patients: a preliminary research. J Exp Clin Cancer Res 32:23
Dührsen U, Müller S, Hertenstein B, Thomssen H, Kotzerke J, Mesters R et al (2018) Positron Emission Tomography-Guided Therapy of Aggressive Non-Hodgkin Lymphomas (PETAL): A multicenter, randomized phase III trial. J Clin Oncol 36:2024–2034
Richter J, Hüttmann A, Rekowski J, Schmitz C, Gärtner S, Rosenwald A et al (2019) Molecular characteristics of diffuse large B-cell lymphoma in Positron Emission Tomography-Guided Therapy of Aggressive Non-Hodgkin lymphomas (PETAL) trial: Correlation with interim PET and outcome. Blood Cancer J 9:67
Rekowski J, Hüttmann A, Schmitz C, Müller SP, Kurch L, Kotzerke J et al (2021) Interim PET evaluation in diffuse large B-cell lymphoma employing published recommendations: comparison of the Deauville 5-point scale and the ΔSUVmax method. J Nucl Med 62:37–42
Schmitz C, Hüttmann A, Müller SP, Hanoun M, Boellaard R, Brinkmann M et al (2020) Dynamic risk assessment based on positron emission tomography scanning in diffuse large B-cell lymphoma: Post-hoc analysis from the PETAL trial. Eur J Cancer 124:25–36
Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G et al (2004) Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 103:275–282
Schwab LP, Peacock DL, Majumdar D, Ingels JF, Jensen LC, Smith KD et al (2012) Hypoxia-inducible factor 1α promotes primary tumor growth and tumor-initiating cell activity in breast cancer. Breast Cancer Res 14:R6
Evens AM, Sehn LH, Farinha P, Nelson BP, Raji A, Lu Y et al (2010) Hypoxia-inducible factor-1α expression predicts superior survival in patients with diffuse large B-cell lymphoma treated with R-CHOP. J Clin Oncol 28:1017–1024
Lirussi L, Antoniali G, D’Ambrosio C, Scaloni A, Nilsen H, Tell G (2016) APE1 polymorphic variants cause persistent genomic stress and affect cancer cell proliferation. Oncotarget 7:26293–26306
Kurland BF, Peterson LM, Shields AT, Lee JH, Byrd DW, Novakova-Jiresova A et al (2019) Test-retest reproducibility of 18 F-FDG PET/CT uptake in cancer patients within a qualified and calibrated local network. J Nucl Med 60:608–614
Acknowledgements
We thank the patients, investigators, and PET centres from all parts of Germany for their participation.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was funded by Deutsche Krebshilfe (Grant Nos. 107592 and 110515 to UD), Amgen Germany, and Roche Pharma (institutional research funding to the University Hospital Essen). The funding bodies had no role in the design of the study, the collection, analysis, and interpretation of data or in writing the manuscript.
Author information
Authors and Affiliations
Contributions
MB-P designed this substudy of the PETAL trial, MB-P and NB-B performed germline genotyping, SPM, HG and AB coordinated the nuclear medicine investigations, AH, CH and UD coordinated the clinical activities, JuRi and WK coordinated the reference pathological review and performed molecular studies on tumour specimens, JaRe and UD performed the statistical analysis, and UD wrote the first draft of the manuscript. All authors contributed to and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
UD: Institutional research grants and honoraria from Amgen Germany and Roche Pharma. The other authors report no potential conflicts of interest relevant to this article.
Ethics approval and consent to participate
The study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of the University of Duisburg-Essen (No. 07-3366). All patients gave written informed consent.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Broecker-Preuss, M., Becher-Boveleth, N., Müller, S.P. et al. Impact of germline polymorphisms in genes regulating glucose uptake on positron emission tomography findings and outcome in diffuse large B-cell lymphoma: results from the PETAL trial. J Cancer Res Clin Oncol 148, 2611–2621 (2022). https://doi.org/10.1007/s00432-021-03796-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-021-03796-z