Abstract
Purpose
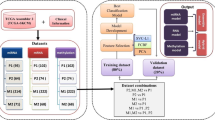
Traditional classification of melanoma is widely utilized with little apparent results making the development of robust classifiers that can guide therapies an urgency. Successful seminal research on classification has provided a wider understanding of cancer from multiple molecular profiles, respectively. However, it may ignore the complementary nature of the information provided by different types of data, which motivated us to subtype melanoma by aggregating multiple genomic platform data.
Methods
Aggregating three omics data of 328 melanoma samples, melanoma subtyping was performed by three clustering methods. Differences across subtypes were extracted by functional enrichment, epigenetically silencing, gene mutations and clinical features. Subtypes were further distinguished by putative biomarkers.
Results
Functional enrichment of the subtype-specific differential expression genes endowed subtypes new designation: immune, melanin and ion, in which the first subtype was enriched for immune system, the second was characterized by melanin and pigmentation, and the third was enriched for ion-involved transmission process. Subtypes also differed in age, Breslow thickness, tumor site, mutation frequency of BRAF, PTGS2, CDKN2A, CDKN2B and incidence of epigenetically silencing for IL15RA, EPSTI1, LXN, CDKN1B genes.
Conclusions
Skin cutaneous melanoma can be robustly divided into three subtypes by SNFCC+. Compared with the TCGA classification derived from gene expression, the subtypes we presented share concordance, but new traits are excavated. Such a genomic classification offers insights to further personalize therapeutic decision-making and melanoma management.




Similar content being viewed by others
Abbreviations
- SKCM:
-
Skin cutaneous melanoma
- TCGA:
-
The Cancer Genome Atlas
- CNV:
-
Copy number variation
- DEG:
-
Differential expression gene
- SNF:
-
Similarity network fusion
- CC:
-
Consensus clustering
- SNFCC+ :
-
Similarity network fusion plus consensus clustering
- HC:
-
Hierarchical clustering
- NMF:
-
Non-negative matrix factorization
- FDR:
-
False discovery rate
- GDC:
-
Genomic data commons
- DAVID:
-
Database for annotation, visualization and integrated discovery
References
Allen D, Lepple-Wienhues A, Cahalan M (1997) Ion channel phenotype of melanoma cell lines. J Membr Biol 155:27–34
Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, Bock C (2014) Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods 11:1138
Austin PF, Cruse CW, Lyman G, Schroer K, Glass F, Reintgen DS (1994) Age as a prognostic factor in the malignant melanoma population. Ann Surg Oncol 1:487–494
Azimi F et al (2012) Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol 30:2678–2683
Balch CM (1992) Cutaneous melanoma: prognosis and treatment results worldwide. Semin Surg Oncol 8:400–414
Balch CM et al (2001) Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. J Clin Oncol 19:3622–3634
Balch CM et al (2009) Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol 27:6199–6206
Bhatia P, Friedlander P, Zakaria EA, Kandil E (2015) Impact of BRAF mutation status in the prognosis of cutaneous melanoma: an area of ongoing research. Ann Transl Med 3:24
Bonafede E (2015) Differential expression analysis for sequence count data via mixtures of negative binomials. Alma
Brunet J-P, Tamayo P, Golub TR, Mesirov JP (2004) Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci 101:4164–4169
Bryois J et al (2014) Cis and trans effects of human genomic variants on gene expression. PLoS Genet 10:e1004461
ChantoôMe AL, Potier-Cartereau M, Roger SB, Vandier C, Soriani O, Joulin V (2013) Ion channels as promising therapeutic targets for melanoma. InTech 20:429–460
Cin H et al (2011) Oncogenic FAM131B–BRAF fusion resulting from 7q34 deletion comprises an alternative mechanism of MAPK pathway activation in pilocytic astrocytoma. Acta Neuropathol 121:763–774
Clark WH, From L, Bernardino EA, Mihm MC (1969) The histogenesis and biologic behavior of primary human malignant melanomas of the skin. Cancer Res 29:705–727
Conteduca G et al (2010) The role of AIRE polymorphisms in melanoma. Clin Immunol 136:96–104
Fecher LA, Cummings SD, Keefe MJ, Alani RM (2007) Toward a molecular classification of melanoma. J Clin Oncol 25:1606–1620
Fisher NM, Schaffer JV, Berwick M, Bolognia JL (2005) Breslow depth of cutaneous melanoma: impact of factors related to surveillance of the skin, including prior skin biopsies and family history of melanoma. J Am Acad Dermatol 53:393–406
Gao J et al (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1
Garg K et al (2016) Tumor-associated B cells in cutaneous primary melanoma and improved clinical outcome. Human Pathol 54:157–164
Gomez-Lira M et al (2014) Association of promoter polymorphism – 765G> C in the PTGS2 gene with malignant melanoma in Italian patients and its correlation to gene expression in dermal fibroblasts. Exp Dermatol 23:766–768
Haluska F, Pemberton T, Ibrahim N, Kalinsky K (2007) The RTK/RAS/BRAF/PI3K pathways in melanoma: biology, small molecule inhibitors, and potential applications. Semin Oncol 34:546–554
Hawkes JE et al (2013) Report of a novel OCA2 gene mutation and an investigation of OCA2 variants on melanoma risk in a familial melanoma pedigree. J Dermatol Sci 69:30–37
Hinoue T et al (2012) Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 22:271–282
Holm K et al (2010) Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns. Breast Cancer Res 12(3):R36
Inamdar GS, Madhunapantula SV, Robertson GP (2010) Targeting the MAPK pathway in melanoma: why some approaches succeed and other fail. Biochem Pharmacol 80:624–637
Innominato PF, Libbrecht L, van den Oord JJ (2001) Expression of neurotrophins and their receptors in pigment cell lesions of the skin. J Pathol 194:95–100
Inozume T et al (2005) Novel melanoma antigen, FCRL/FREB, identified by cDNA profile comparison using DNA chip are immunogenic in multiple melanoma patients. Int J Cancer 114:283–290
Jacquelot N et al (2016) Chemokine receptor patterns in lymphocytes mirror metastatic spreading in melanoma. J Clin Investig 126:921–937
Jayakumar A, Rauvolfova J, Bao H, Fokt I, Skora S, Heimberger A, Priebe W (2013) Abstract 3251: Blockade of HIF-1 with a small molecule inhibitor WP1066 in melanoma. Cancer Res 73:3251–3251
Jemal A, Siegel R, Xu J, Ward E (2010) Cancer statistics, 2010 CA: a cancer. J Clin 60:277–300
Jonsson G et al (2010) Gene expression profiling-based identification of molecular subtypes in stage IV Melanomas with different clinical outcome clinical cancer research an official. J Am Assoc Cancer Res 16:3356–3367
Journe F et al (2011) TYRP1 mRNA expression in melanoma metastases correlates with clinical outcome. Br J Cancer 105:1726–1732
Kawaguchi M, Hearing VJ (2011) The roles of ADAMs family proteinases in skin diseases. Enzyme Res 2011:482498
Koh SS et al (2012) Differential gene expression profiling of primary cutaneous melanoma and sentinel lymph node metastases. Mod Pathol 25:828–837
Li M, Xiong Z-G (2011) Ion channels as targets for cancer therapy. Int J Physiol Pathophysiol Pharmacol 3:156–166
Li T et al (2013) Identification of epithelial stromal interaction 1 as a novel effector downstream of Krüppel-like factor 8 in breast cancer invasion and metastasis. Oncogene 33:4746–4755
Li FJ et al (2014) Emerging Roles for the FCRL Family Members in Lymphocyte Biology and Disease. Curr Top Microbiol Immunol 382:29–50
Liu W, Peng Y, Tobin DJ (2013) A new 12-gene diagnostic biomarker signature of melanoma revealed by integrated microarray analysis PeerJ 1:e49
Lodolce J et al (2002) Interleukin-15 and the regulation of lymphoid homeostasis. Mol Immunol 39:537–544
Macià A, Herreros J, Martí RM, Cantí C (2015) Calcium channel expression and applicability as targeted therapies in melanoma. Biomed Res Int 2015:587135
Maurer M, Somasundaram R, Herlyn M, Wagner SN (2012) Immunotargeting of tumor subpopulations in melanoma patients: A paradigm shift in therapy approaches. Oncoimmunology 1:1454–1456
McGovern VJ et al (1973) The classification of malignant melanoma and its histologic reporting. Cancer 32:1446–1457
Monti S, Tamayo P, Mesirov J, Golub T (2003) Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Mach Learn 52:91–118
Palmieri G (2012) Molecular classification of patients with cutaneous melanoma: a reality. J Mol Biomark Diagn 3:e110
Payne AS, Cornelius LA (2002) The role of chemokines in melanoma tumor growth and metastasis. J Investig Dermatol 118:915–922
Rogers HW, Weinstock MA, Feldman SR, Coldiron BM (2015) Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the US Population, 2012. Jama Dermatol 151:1081–1086
Rousseeuw PJ (1987) Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20:53–65
Scolyer RA, Long GV, Thompson JF (2011) Evolving concepts in melanoma classification and their relevance to multidisciplinary melanoma patient care. Mol Oncol 5:124–136
Slominski R, Zmijewski M, Slominski AT (2015) On the role of melanin pigment in melanoma. Exp Dermatol 24:258–259
Stern RS (2010) Prevalence of a history of skin cancer in 2007: results of an incidence-based model. Arch Dermatol 146:279–282
TCGA (2015) Genomic classification of cutaneous melanoma. Cell 161:1681–1696
Verreault M, Webb MS, Ramsay EC, Bally MB (2006) Gene silencing in the development of personalized cancer treatment: the targets, the agents and the delivery systems. Curr Gene Ther 6:505–533
Wang B et al (2014) Similarity network fusion for aggregating data types on a genomic scale. Nat Methods 11:333–337
Xu T et al (2017) CancerSubtypes: an R/Bioconductor package for molecular cancer subtype identification, validation, and visualization. Bioinformatics 33:3131–3133
Zhao Q, Shi X, Xie Y, Huang J, Shia B, Ma S (2015) Combining multidimensional genomic measurements for predicting cancer prognosis: observations from TCGA. Brief Bioinform 16:291–303
Funding
This study was funded by the National Social Science Fund under award No. 16BTJ021.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Availability of data and material
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request and the data sets are also available in the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lu, X., Zhang, Q., Wang, Y. et al. Molecular classification and subtype-specific characterization of skin cutaneous melanoma by aggregating multiple genomic platform data. J Cancer Res Clin Oncol 144, 1635–1647 (2018). https://doi.org/10.1007/s00432-018-2684-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-018-2684-7