Abstract
Type IV pili are versatile and highly flexible fibers formed on the surface of many Gram-negative and Gram-positive bacteria. Virulence and infection rate of several pathogenic bacteria, such as Neisseria meningitidis and Pseudomonas aeruginosa, are strongly dependent on the presence of pili as they facilitate the adhesion of the bacteria to the host cell. Disruption of the interactions between the pili and the host cells by targeting proteins involved in this interaction could, therefore, be a treatment strategy. A type IV pilus is primarily composed of multiple copies of protein subunits called major pilins. Additional proteins, called minor pilins, are present in lower abundance, but are essential for the assembly of the pilus or for its specific functions. One class of minor pilins is required to initiate the formation of pili, and may form a complex similar to that identified in the related type II secretion system. Other, species-specific minor pilins in the type IV pilus system have been shown to promote additional functions such as DNA binding, aggregation and adherence. Here, we will review the structure and the function of the minor pilins from type IV pili.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Motility and pathogenicity of various bacteria are strongly dependent on proteinaceous appendages called pili, located at the cell surface [1, 2]. Pili are highly flexible, long fibers assembled in the envelope of bacteria. Several different types of pili have been identified and grouped based on their structure and assembly mechanism [3]. Both Gram-negative and Gram-positive bacteria can form pili, though the diversity of pili formed in Gram-negative bacteria is larger [3, 4]. In this review, we will focus on the type IV pili (T4P), which promote biological functions important for the pathogenicity of bacteria such as twitching motility, DNA uptake and adhesion to host cells [1]. The T4P family is generally subdivided into two groups, type IVa pili (T4aP) and type IVb pili (T4bP). This subdivision is based on the sequence of the pilins, the length of the leader peptide, and minor differences in their assembly [1, 5]. The organization of the genes required for pilus formation are different between T4aP and T4bP systems. T4aP genes are spread throughout the genome in several operons, whereas the T4bP genes are clustered in a single operon [2]. T4bP are involved in biofilm formation, bacterial colonization and cell adhesion [6]. T4aP have been identified as the molecular framework behind bacterial twitching motility and other functions such as DNA uptake [7]. Their flexibility, elongation and retraction allow them to ensure these different functions.
This review will highlight recent advances in the understanding of the assembly of T4P as well as in the biological functions and the structure of their minor pilins. Although the T4P assembly system is conserved between bacterial species, the nomenclature of T4P proteins is very heterogeneous. In this review, unless stated otherwise, we will use the nomenclature from Pseudomonas aeruginosa, one of the best-characterized T4P systems (Table 1).
Building blocks of a pilus: the pilins
The major building blocks of a T4P are proteins called pilins, which are composed of a periplasmic domain and a transmembrane helix [3] (Fig. 1a). The bulk of a pilus is made up of thousands of copies of one subunit called the major pilin (Fig. 1b). Other pilins with a lower abundance, the minor pilins, have been shown to be essential for pilus formation and function [8]. Minor pilins can be further categorized into core and non-core minor pilins [9]. Core minor pilins are important for the formation of pili, despite their low abundance.
A major pilin and the type IVa pilus. a Molecular representation of the P. aeruginosa T4aP major pilin PilA (PDB:1OQW). The central α-helix is colored in red, the transmembrane N-terminal part (α1-N) and C-terminal (α1-C) part of the folded domain are indicated. The αβ-loop and D-region are highlighted in green and magenta, respectively. b The surface representation of the pilus assembled by subunits of PilA colored in gray and the αβ-loop and D-region are colored green and magenta, respectively (PDB: 5VXY). The zoom-in highlights the surface exposed of one PilA subunit in the pilus and the interactions between neighboring pilins
Pilins, both major and minor, are produced as prepilins with a short N-terminal leader sequence (signal peptide), which serves as an anchor to ensure correct insertion of the pilins in the cytoplasmic membrane [10]. The SecYEG translocon promotes prepilin membrane insertion in such a way that the N-terminal peptide is cytosolically exposed and the C-terminal domain of the pilins is exported to the periplasm (Fig. 1a) [12, 13]. The leader peptide is cleaved off by a specific prepilin peptidase to obtain the mature pilins that are competent for assembly [11].
The structural features of T4a pilins, both major and minor, are very similar (see Figs. 1a, 2a, b). They all adopt a lollipop-like shape with an N-terminal helix (α1), followed by a globular domain [9]. The α1 helix can further be separated into two segments, a hydrophobic N-terminal transmembrane (α1-N) and a C-terminal (α1-C) parts, which extends into the globular domain. Before assembly into the pilus, free pilins are anchored in the inner membrane by the transmembrane part (α1-N). At the C-terminal end of the transmembrane domain, the sequence contains a stretch of conserved residues usually limited by a glycine and a proline. In all known structures of T4P and the highly homologous type II secretion system (T2SS) major pilin subunits in the context of the pilus, this stretch disrupts the helix and forms an extended structure when incorporated into the fiber [14,15,16,17]. In the pilus, this region is also accessible to solvent as assessed by hydrogen–deuterium exchange in a T2SS pseudopilus [18] and a T4b pilus [19]. This conserved structural feature may enable the pilus to undergo spring-like extensions in response to shear forces [17]. Nothing is known about the structure of this region in minor pilins, since structural studies of individual minor pilins were limited to the soluble globular domains, and no structure of a complete pilus with a minor pilin is known.
Comparison of minor pilins of T4aP from P. aeruginosa (or equivalent); the α1-helix (red), αβ-loop (green) and D-region (magenta) are highlighted. Disulphide bonds are represented as sticks. a Structures of core minor pilins of P. aeruginosa or equivalent pilins minor from T. thermophilus. We include these structures although the assignment as core minor pilins is indirect, by homology to GspI and GspJ. b Structures of non-core minor pilins of P. aeruginosa or equivalent minor pilins from N. meningitis. c Complex of three core minor pseudopilins of enterotoxigenic E. coli, GspI-J-K. GspI is colored in blue, GspJ is colored in green, the pilin domain of GspK is colored in brown and the inserted domain of GspK is colored in gray. An area covered by this inserted domain, which would form a solvent accessible cavity otherwise, is highlighted. Figure was prepared with The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC
The central axis of the globular domain is formed by the α1-C helix, which is surrounded by one or two antiparallel β-sheets composed of 4–7 strands, slightly rotated compared to the α-helical axis (Figs. 1a, 2a, b). Major structural differences between different minor and major pilins are restricted to the characteristic hypervariable regions: the αβ-loop connecting the α1-C helix to the β-sheet and the D-region enclosed by the C-terminal disulfide bridge found in some species. The αβ-loop is structurally very versatile and can contain various structural motifs. The ɑβ-loop of FimU includes distorted β-strands, whereas the D-region contains two antiparallel β-strands (Fig. 2a) [20]. Remarkably, the equivalents of PilV and PilW from Thermus thermophilus, Tt1218 and Tt1219, respectively, do not have a D-region, as the mostly conserved C-terminal disulfide bridge is absent in both proteins [21]. The C-terminal β-sheet has the same elongated shape in both pilins, the major difference is restricted to the αβ-loop. Tt1218 has a very short ɑβ-loop composed of only a short α-helix and a single β-strand. The ɑβ-loop of Tt1219 is much longer and contains a 5-stranded antiparallel β-sheet and two disulfide bridges.
Assembly, elongation and retraction of pili
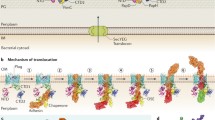
The T4P assembly system is responsible for the elongation and the retraction of pili. In Gram-negative bacteria, the T4P assembly system is composed of between 10 and 18 different proteins forming complexes that are located in the inner membrane, in the periplasm and in the outer membrane [2]. Recent studies of the T4aP from Myxococcus xanthus [22] and T. thermophilus [23] by electron cryo-tomography have revealed the in situ organization of key elements in this assembly system that are summarized below (Fig. 3a) [22].
Type IVa pilus (T4aP) assembly. a The T4aP assembly machinery from M. xanthus (PDB:3JC8). On the left the outer membrane (OM), the periplasmic space (PS) and the inner membrane (IM) are marked. The individual proteins involved in the formation of T4P are shown and labeled. On the right the spanning of the different sub-complexes (pilus, OM pore, alignment complex, pilus assembly platform and motor) are indicated, see text for detailed description. b Schematic overview of the pilus initiation and elongation of tip complex. The orientations of the minor pilins are shown as hypothesized
Elongation and retraction of the pilus are made possible by the addition or removal, respectively, of major pilin subunits (PilA) at the base of the pilus, and are facilitated by the central inner membrane platform protein, PilC and two antagonistic ATPase motors, PilB and PilT (Fig. 3) [24, 25]. PilC is in direct contact with the pilins and with the ATPases, which presumably rotate PilC to either elongate or retract the pilus [26, 27]. PilB and PilT occupy the center of the PilM cytoplasmic ring positioned near the inner membrane [28]. The PilM cytoplasmic ring interacts with the N-terminal transmembrane tail of PilN [28]. PilN forms a heterodimer with PilO, which assembles as a ring structure in the periplasm with the same stoichiometry as the cytoplasmic ring. The inner membrane protein PilP binds to this complex and connects it to the periplasmic domain of PilQ. The large PilQ multimer forms the secretin pore in the outer membrane necessary for the passage of the pilus through the membrane [29, 30]. Thus, the PilMNOP subcomplex links the cytoplasmic ATPases to the secretin pore [22, 31,32,33] and ensures that the motors of the assembly platform, PilC and PilB/PilT, are aligned with the secretin [34,35,36]. The fibers built by inserting pilins into the pilus base extend through the secretin pore out of the cell to perform their biological functions.
The T4P machinery is highly homologous to the type II secretion system (T2SS), in both assembly mechanism and pilus structure [37]. The sequence and structural organization of pilins of the two systems are conserved, and a T4P can be efficiently assembled by the T2SS [38]. The pilus, or the pseudopilus in T2SS, allows specific folded proteins in the periplasm to be secreted to the external medium. In T2SS, the minor pseudopilins promote assembly initiation [39] and have been implicated in substrate binding [40]. The absence of a retraction ATPase in the T2SS assembly platform is one of the major differences with the T4P system. The importance of calcium for the stability of the pseudopili from Klebsiella oxytoca suggests its implication in length control and passive disassembly of the pilus [14].
Structure and function of core minor pilins
In the T4aP system, there are typically four core minor pilin genes that are clustered in an operon, which are homologous to the genes encoding four minor pseudopilins in the T2SS (Table 1). Deletion of these genes results in strong defects in pilus assembly and function, as demonstrated in P. aeruginosa [20] and enterohemorrhagic Escherichia coli (EHEC) [41]. In N. meninigitidis, deletion of each of the minor pilin genes—pilH, I, J and K—leads to a non-piliated phenotype [42]. Mutation in the ATPase pilT restores piliation, which led to the suggestion [43] that the role of these minor pilins is “anti-retraction”. However, each of them is required for fully efficient piliation, by analogy to the role of minor pseudopilins in the T2SS, suggesting that they are core minor pilins. A detailed study of the role of individual minor pilins in piliation and twitching motility demonstrated the role of the core minor pilins in assembly initiation, and showed that in the pilT mutant background, the residual piliation is dependent on minor T2SS pseudopilins [8, 44]. This confirms that T4a and T2SS minor pilins are functionally interchangeable, as had been shown before [45].
The homology between the four core minor pilins in T4P and T2SS led to the suggestion that they interact and form a complex responsible for the initiation of the pilus assembly in similar ways in both systems (Fig. 2c). The X-ray crystal structure of a complex of three minor pseudopilins, GspI, GspJ and GspK, from the T2SS of enterotoxigenic E. coli (ETEC) [46] serves as a structural template. In this complex, the tip subunit GspK acts as a cap, strongly suggesting that the core minor pilins are assembled first to form the tip of the nascent pseudopilus [46]. The minor pseudopilins form a staggered complex in the membrane, promoting initiation and setting the register for helical pseudopilus assembly [39].
This minor pilin complex would stabilize the tip of the pilus and provide a template for assembly of major subunits (Fig. 3b). In analogy to the T2SS, some of the minor pilins should differ both in sequence and structure from the major pilin to form a stable complex, which can remain anchored at the pilus tip. The GspI-J-K complex is composed of pilins of different molecular weight and three-dimensional shape (Fig. 2c), the central GspI being the smallest and GspK the largest, more than twice the size of the major pilin GspG. GspK has a large insertion in the globular domain, which might ensure that the complex is only incorporated at the tip, and its increased size can be imagined to shield the cavity that is formed by pilins of similar size and structure at the tip of a pilus (Fig. 2c). Unfortunately, there is no structure available for the GspK equivalent PilX from P. aeruginosa, nor for an equivalent T4P pilin from other species.
Although the sequence similarity makes it likely that a similar tip complex exists also in the T4P system, no structural information of this type of complex has been obtained to this date in the T4aP system. Remarkably, the large insertion present in GspK is absent in the T4P equivalent PilX from P. aeruginosa, and one has to postulate a different mechanism to cap the pilus. For example, in the T4bP from ETEC a single minor subunit CofB promotes initiation of assembly. Structural studies revealed that this subunit contains four distinct domains corresponding those of T4aP core minor pilins [47]. An interesting model based on another crystal structure proposes that CofB forms a homo-trimer that caps and primes pilus assembly [48]. Its homologue TcpB, the unique minor pilin in the Vibrio cholerae T4bP has been implicated in initiation of pilus assembly and disassembly [49]. Furthermore, recent biochemical evidence suggest that it forms a trimer localized at the pilus tip [50].
Structure and function of non-core minor pilins
The non-core minor pilins have a more conserved fold and the diversity in the αβ-loop, and the D-region is more limited compared to the core minor pilins (see Fig. 2b). The αβ-loops of PilE [51] and the equivalents from N. meningitidis PilX [52] and PilV, are shorter and contain a small α-helical segment. The D-region in PilE and PilV from N. meningitidis both contain two β-strands, which are part of the conserved C-terminal β-sheet. Furthermore, PilE and PilX from N. meningitidis have a short helical segment in the D-region.
Non-core minor pilins are involved in the different biological functions that are promoted by T4P, including aggregation, adhesion and natural competence—the acquisition of external DNA (either inter or intra species) [7]. In Neisseria species, the minor pilin PilX promotes aggregation via pilus–pilus interactions [52], whereas PilV promotes adherence and signaling via receptors on eukaryotic cell surface [53]. In some species, additional proteins affect T4P-mediated adhesion. For example, PilY1 from P. aeruginosa is an adhesin which interacts with the complex formed by core minor pilins at the tip of the pilus [20]. PilY1 could also facilitate the adhesion to host epithelial cells. In addition, PilY1 and other minor pilins are involved in the regulation of P. aeruginosa virulence genes [54]. In EHEC no pilin-associated adhesins have been identified, but the expression of T4aP genes promotes twitching motility [55].
The minor pilin of N. meningitidis ComP binds DNA, recognizing a specific DNA sequence called the uptake signal [56] to promote natural transformation [57]. The DNA uptake mechanism mediated by T4P is not completely understood. However, it has been proposed that T4P play a role in the early step of DNA uptake [7]. A study of competence T4P from Vibrio cholerae by live fluorescence microscopy showed that DNA binds at the tip of the pilus [58]. This suggests that the binding of DNA is happening at the pilus tip principally via the minor pilins, despite their low abundance. ComP from the Neisseria species is also thought to be located at the pilus tip [59]. The retraction of the T4P brings the DNA to the cell surface where the DNA can be internalized via the secretin pore of the pilus or an alternate channel [7].
Some non-core minor pilins have large structural insertions. The recently determined structure of the minor T4a pilin ComZ from T. thermophilus [60] shows an unusual structure with two domains, a pilin-like domain and a large additional domain inserted into its β-sheet. Hence, although not involved in pilus assembly, this minor pilin is likely located at the tip of the pilus, since the structural organization and the large size of this additional domain cannot allow upward incorporation of major pilins. Interestingly, this domain of ComZ was shown to be involved in DNA binding.
Concluding remarks
With the emergence of antibiotic resistance, T4P are considered as a target for innovative antibacterial therapeutics [61]. T4aP of EHEC, among others, have been shown to be involved in biofilm formation, twitching motility and adherence [60]. The core minor pilins that likely form the tip of the pilus might play multiple roles, being at the same time responsible for the initiation of pilus formation, capping the pilus, and mediate specific biological functions. However, it is at the moment not clear if only the tip complex or also the major pilins can interact with surfaces or host cells. Further work is thus required in order to resolve the specific structural and biological mechanism of T4aP. Detailed structural knowledge of the minor pilins in the context of the entire fiber would be a major asset for the development of new vaccinal and therapeutic strategies.
Abbreviations
- T2SS:
-
Type II secretion system
- T4P:
-
Type IV pilus
- T4aP:
-
Type IVa pilus
- T4bP:
-
Type IVb pilus
References
Craig L, Pique ME, Tainer JA (2004) Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol 2:363–378
Pelicic V (2008) Type IV pili: e pluribus unum? Mol Microbiol 68:827–837. https://doi.org/10.1111/j.1365-2958.2008.06197.x
Hospenthal MK, Costa TRD, Waksman G (2017) A comprehensive guide to pilus biogenesis in Gram-negative bacteria. Nat Rev Microbiol 15:365–379
Van Gerven N, Waksman G, Remaut H (2011) Pili and flagella: biology, structure, and biotechnological applications. Academic, Oxford, pp 21–72
Ayers M, Howell PL, Burrows LL (2010) Architecture of the type II secretion and type IV pilus machineries. Future Microbiol 5:1203–1218
Roux N, Spagnolo J, De Bentzmann S (2012) Neglected but amazingly diverse type IVb pili. Res Microbiol 163:659–673. https://doi.org/10.1016/j.resmic.2012.10.015
Piepenbrink KH (2019) DNA uptake by type IV filaments. Front Mol Biosci 6:1–13. https://doi.org/10.3389/fmolb.2019.00001
Giltner CL, Habash M, Burrows LL (2010) Pseudomonas aeruginosa minor pilins are incorporated into type IV Pili. J Mol Biol 398:444–461. https://doi.org/10.1016/j.jmb.2010.03.028
Giltner CL, Nguyen Y, Burrows LL (2012) Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev 76:740–772. https://doi.org/10.1128/MMBR.00035-12
Berry JL, Pelicic V (2015) Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS Microbiol Rev 39:134–154
Strom MS, Nunn DN, Lory S (1993) A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Natl Acad Sci USA 90:2404–2408
Arts J, van Boxtel R, Filloux A, Tommassen J, Koster M (2007) Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle-Sec pathway. J Bacteriol 189:2069–2076
Francetic O, Buddelmeijer N, Lewenza S, Kumamoto CA, Pugsley AP (2007) Signal recognition particle-dependent inner membrane targeting of the PulG Pseudopilin component of a type II secretion system. J Bacteriol 189:1783–1793
López-Castilla A, Thomassin JL, Bardiaux B, Zheng W, Nivaskumar M, Yu X, Nilges M, Egelman EH, Izadi-Pruneyre N, Francetic O (2017) Structure of the calcium-dependent type 2 secretion pseudopilus. Nat Microbiol 2:1686–1695. https://doi.org/10.1038/s41564-017-0041-2
Bardiaux B, de Amorim GC, Luna Rico A, Zheng W, Guilvout I, Jollivet C, Nilges M, Egelman EH, Izadi-Pruneyre N, Francetic O (2019) Structure and assembly of the enterohemorrhagic Escherichia coli type 4 pilus. Structure. https://doi.org/10.1016/j.str.2019.03.021
Wang F, Coureuil M, Osinski T, Orlova A, Altindal T, Gesbert G, Nassif X, Egelman EH, Craig L (2017) Cryoelectron microscopy reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae Type IV Pili at sub-nanometer resolution. Structure 25:1423–1435.e4. https://doi.org/10.1016/j.str.2017.07.016
Kolappan S, Coureuil M, Yu X, Nassif X, Egelman EH, Craig L (2016) Structure of the neisseria meningitidis type IV pilus. Nat Commun 7:1–12. https://doi.org/10.1038/ncomms13015
Bardiaux B, Cordier F, Brier S, López-Castilla A, Izadi-Pruneyre N, Nilges M (2019) Dynamics of a type 2 secretion system pseudopilus unraveled by complementary approaches. J Biomol NMR. 73(6–7):293–303. https://doi.org/10.1007/s10858-019-00246-4(Epub 2019 May 23)
Li J, Lim MS, Li S, Brock M, Pique ME, Woods VL Jr, Craig L (2008) Vibrio cholerae toxin-coregulated pilus structure analyzed by hydrogen/deuterium exchange mass spectrometry. Structure. 16(1):137–148. https://doi.org/10.1016/j.str.2007.10.027
Sugiman-Marangos S, Junop MS, Nguyen Y, Bell SD, Burrows LL, Harvey H, Charlton CL (2014) Pseudomonas aeruginosa minor pilins prime Type IVa pilus assembly and promote surface display of the PilY1 adhesin. J Biol Chem 290:601–611. https://doi.org/10.1074/jbc.m114.616904
Karuppiah V, Thistlethwaite A, Derrick JP (2016) Structures of type IV pilins from Thermus thermophilus demonstrate similarities with type II secretion system pseudopilins. J Struct Biol 196:375–384. https://doi.org/10.1016/j.jsb.2016.08.006
Chang Y-W, Rettberg LA, Treuner-Lange A, Iwasa J, Søgaard-Andersen L, Jensen GJ (2016) Architecture of the type IVa pilus machine. Science 351:aad2001. https://doi.org/10.1126/science.aad2001
Gold VA, Salzer R, Averhoff B, Kühlbrandt W (2015) Structure of a type IV pilus machinery in the open and closed state. Elife. 21:4. https://doi.org/10.7554/eLife.07380
Takhar HK, Kemp K, Kim M, Howell PL, Burrows LL (2013) The platform protein is essential for type IV pilus biogenesis. J Biol Chem 288:9721–9728. https://doi.org/10.1074/jbc.M113.453506
Tsai CL, Tainer JA (2016) The ATPase motor turns for type IV pilus assembly. Structure 24:1857–1859
Mancl JM, Black WP, Robinson H, Yang Z, Schubot FD (2016) Crystal structure of a type IV pilus assembly ATPase: insights into the molecular mechanism of PilB from Thermus thermophilus. Structure 24:1886–1897. https://doi.org/10.1016/j.str.2016.08.010
Georgiadou M, Castagnini M, Karimova G, Ladant D, Pelicic V (2012) Large-scale study of the interactions between proteins involved in type IV pilus biology in Neisseria meningitidis: characterization of a subcomplex involved in pilus assembly. Mol Microbiol 84:857–873. https://doi.org/10.1111/j.1365-2958.2012.08062.x
Karuppiah V, Derrick JP (2011) Structure of the PilM-PilN inner membrane type IV pilus biogenesis complex from Thermus thermophilus. J Biol Chem 286:24434–24442. https://doi.org/10.1074/jbc.M111.243535
Berry JL, Phelan MM, Collins RF, Adomavicius T, Tønjum T, Frye SA, Bird L, Owens R, Ford RC, Lian LY, Derrick JP (2012) Structure and assembly of a trans-periplasmic channel for type IV pili in Neisseria meningitidis. PLoS Pathog. https://doi.org/10.1371/journal.ppat.1002923
Wolfgang M, Putten JPM, Hayes S, Dorward D, Koomey M (2000) Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J 19:6408–6418. https://doi.org/10.1093/emboj/19.23.6408
Carter T, Buensuceso RNC, Tammam S, Lamers RP, Harvey H, Howell PL, Burrows LL (2017) The type IVa pilus machinery is recruited to sites of future cell division. MBio. https://doi.org/10.1128/mBio.02103-16
Siewering K, Jain S, Friedrich C, Webber-Birungi MT, Semchonok DA, Binzen I, Wagner A, Huntley S, Kahnt J, Klingl A, Boekema EJ, Søgaard-Andersen L, van der Does C (2014) Peptidoglycan-binding protein TsaP functions in surface assembly of type IV pili. Proc Natl Acad Sci 111:E953–E961. https://doi.org/10.1073/pnas.1322889111
Burrows LL, Sundaram P, Koo J, Andrew Chong P, Howell PL, Sampaleanu LM, Tammam S, Ayers M, Forman-Kay JD (2011) Characterization of the PilN, PilO and PilP type IVa pilus subcomplex. Mol Microbiol 82:1496–1514. https://doi.org/10.1111/j.1365-2958.2011.07903.x
Tammam S, Sampaleanu LM, Koo J, Manoharan K, Daubaras M, Burrows LL, Howell PL (2013) PilMNOPQ from the Pseudomonas aeruginosa type IV pilus system form a transenvelope protein interaction network that interacts with PilA. J Bacteriol 195(10):2126–2135. https://doi.org/10.1128/jb.00032-13(Epub 2013 Mar 1)
Balasingham SV, Collins RF, Assalkhou R, Homberset H, Frye SA, Derrick JP, Tønjum T (2007) Interactions between the lipoprotein PilP and the secretin PilQ in Neisseria meningitidis. J Bacteriol. 189(15):5716–5727 (Epub 2007 May 25)
Drake SL, Sandstedt SA, Koomey M (1997) PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass multimer. Mol Microbiol 23(4):657–668
Thomassin JL, Santos Moreno J, Guilvout I, Tran Van Nhieu G, Francetic O (2017) The trans-envelope architecture and function of the type 2 secretion system: new insights raising new questions. Mol Microbiol 105:211–226. https://doi.org/10.1111/mmi.13704
Sauvonnet N, Vignon G, Pugsley AP, Gounon P (2000) Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J 19:2221–2228
Cisneros DA, Bond PJ, Pugsley AP, Campos M, Francetic O (2012) Minor pseudopilin self-assembly primes type II secretion pseudopilus elongation. EMBO J 31:1041–1053. https://doi.org/10.1038/emboj.2011.454
Douzi B, Ball G, Cambillau C, Tegoni M, Voulhoux R (2011) Deciphering the Xcp Pseudomonas aeruginosa type II secretion machinery through multiple interactions with substrates. J Biol Chem 286:40792–40801. https://doi.org/10.1074/jbc.M111.294843
Luna Rico A, Zheng W, Petiot N, Egelman EH, Francetic O (2019) Functional reconstitution of the type IVa pilus assembly system from enterohaemorrhagic Escherichia coli. Mol Microbiol 111:732–749. https://doi.org/10.1111/mmi.14188
Carbonnelle E, Hélaine S, Prouvensier L, Nassif X, Pelicic V (2005) Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol Microbiol 55(1):54–64
Carbonnelle E, Helaine S, Nassif X, Pelicic V (2006) A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol Microbiol 61(6):1510–1522
Nguyen Y, Sugiman-Marangos S, Harvey H, Bell SD, Charlton CL, Junop MS, Burrows LL (2015) Pseudomonas aeruginosa minor pilins prime type IVa pilus assembly and promote surface display of the PilY1 adhesin. J Biol Chem. 290(1):601–611
Cisneros DA, Pehau-Arnaudet G, Francetic O (2012) The heterologous assembly of type IV pilin by a type II secretion system reveals the role of minor pilins in assembly initiation. Mol Microbiol 86(4):805–818
Korotkov KV, Hol WGJ (2008) Structure of the GspK-GspI-GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system. Nat Struct Mol Biol 15:462–468. https://doi.org/10.1038/nsmb.1426
Kolappan S, Ng D, Yang G, Harn T, Craig L (2015) Crystal structure of the minor pilin CofB, the initiator of CFA/III pilus assembly in enterotoxigenic Escherichia coli. J Biol Chem 290:25805–25818. https://doi.org/10.1074/jbc.M115.676106
Kawahara K, Oki H, Fukakusa S, Yoshida T, Imai T, Maruno T, Kobayashi Y, Motooka D, Iida T, Ohkubo T, Nakamura S (2016) Homo-trimeric structure of the type IVb minor pilin CofB suggests mechanism of CFA/III pilus assembly in human enterotoxigenic Escherichia coli. J Mol Biol 428:1209–1226. https://doi.org/10.1016/j.jmb.2016.02.003
Ng D, Harn T, Altindal T, Kolappan S, Marles JM, Lala R, Spielman I, Gao Y, Hauke CA, Kovacikova G, Verjee Z, Taylor RK, Biais N, Craig L (2016) The vibrio cholerae minor pilin TcpB initiates assembly and retraction of the toxin-coregulated pilus. PLoS Pathog 12(12):e1006109. https://doi.org/10.1371/journal.ppat.1006109
Gutierrez-Rodarte M, Kolappan S, Burrell BA, Craig L (2019) The Vibrio cholerae minor pilin TcpB mediates uptake of the cholera toxin phage CTXφ. J Biol Chem 294:15698–15710
Helaine S, Dyer DH, Nassif X, Pelicic V, Forest KT (2007) 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili. Proc Natl Acad Sci 104:15888–15893. https://doi.org/10.1073/pnas.0707581104
Nguyen Y, Harvey H, Sugiman-Marangos S, Bell SD, Buensuceso RNC, Junop MS, Burrows LL (2015) Structural and functional studies of the Pseudomonas aeruginosa Minor Pilin, PilE. J Biol Chem 290:26856–26865. https://doi.org/10.1074/jbc.M115.683334
Bernard SC, Simpson N, Join-Lambert O, Federici C, Laran-Chich MP, Maïssa N, Bouzinba-Ségard H, Morand PC, Chretien F, Taouji S, Chevet E, Janel S, Lafont F, Coureuil M, Segura A, Niedergang F, Marullo S, Couraud PO, Nassif X, Bourdoulous S (2014) Pathogenic Neisseria meningitidis utilizes CD147 for vascular colonization. Nat Med 20:725–731. https://doi.org/10.1038/nm.3563
Marko VA, Kilmury SLN, MacNeil LT, Burrows LL (2018) Pseudomonas aeruginosa type IV minor pilins and PilY1 regulate virulence by modulating FimS-AlgR activity. PLoS Pathog. https://doi.org/10.1371/journal.ppat.1007074
Saldana Z, Giron JA, Monteiro-Neto V, Ledesma MA, Xicohtencatl-Cortes J, Puente JL, Monteiro-Neto V, Saldaña Z, Ledesma MA, Puente JL, Girón JA (2008) The type 4 pili of enterohemorrhagic Escherichia coli O157:H7 are multipurpose structures with pathogenic attributes. J Bacteriol 91:411–421. https://doi.org/10.1128/jb.01306-08
Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, Brady J, Baldwin GS, Lea SM, Matthews SJ, Pelicic V (2013) Specific DNA recognition mediated by a type IV pilin. Proc Natl Acad Sci 110:3065–3070. https://doi.org/10.1073/pnas.1218832110
Wolfgang M, Van Putten JPM, Hayes SF, Koomey M (1999) The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol Microbiol 31:1345–1357. https://doi.org/10.1046/j.1365-2958.1999.01269.x
Ellison CK, Dalia TN, Vidal Ceballos A, Wang JCY, Biais N, Brun YV, Dalia AB (2018) Retraction of DNA-bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae. Nat Microbiol 3:773–780. https://doi.org/10.1038/s41564-018-0174-y
Aas FE, Løvold C, Koomey M (2002) An inhibitor of DNA binding and uptake events dictates the proficiency of genetic transformation in Neisseria gonorrhoeae: mechanism of action and links to Type IV pilus expression. Mol Microbiol 46:1441–1450. https://doi.org/10.1046/j.1365-2958.2002.03265.x
Salleh MZ, Karuppiah V, Snee M, Thistlethwaite A, Levy CW, Knight D, Derrick JP (2019) Structure and properties of a natural competence-associated pilin suggest a unique pilus tip-associated DNA receptor. MBio 10:1–17. https://doi.org/10.1128/mBio
Duménil G (2019) Type IV pili as a therapeutic target. Trends Microbiol 27:658–661
Acknowledgements
We are grateful to the Institut Pasteur and the CNRS for their continued support for our research. This work was funded by the French Agence Nationale de la Recherche (ANR-14-CE09-0004 to OF), the INCEPTION project (PIA/ANR-16-CONV-0005), the FRM (Equipe FRM 2017 M.DEQ 20170839114 to MN) and the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Sklodowska-Curie Grant agreement 765042.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by Volkhard A.J. Kempf.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Jacobsen, T., Bardiaux, B., Francetic, O. et al. Structure and function of minor pilins of type IV pili. Med Microbiol Immunol 209, 301–308 (2020). https://doi.org/10.1007/s00430-019-00642-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00430-019-00642-5