Abstract
Influenza remains an important threat for human health, despite the extensive study of influenza viruses and the production of effective vaccines. In contrast to virus genetics determinants, host genetic factors with clinical impact remained unexplored until recently. The association between three single nucleotide polymorphisms (SNPs) and influenza outcome in a European population was investigated in the present study. All samples were collected during the influenza A(H1N1)pdm09 post-pandemic period 2010–11 and a sufficient number of severe and fatal cases was included. Host genomic DNA was isolated from pharyngeal samples of 110 patients from northern Greece with severe (n = 59) or mild (n = 51) influenza A(H1N1)pdm09 disease, at baseline, and the genotype of CD55 rs2564978, C1QBP rs3786054 and FCGR2A rs1801274 SNPs was investigated. Our findings suggest a relationship between the two complement-related SNPs, namely, the rare TT genotype of CD55 and the rare AA genotype of C1QBP with increased death risk. No significant differences were observed for FCGR2A genotypes neither with fatality nor disease severity. Additional large-scale genetic association studies are necessary for the identification of reliable host genetic risk factors associated with influenza A(H1N1)pdm09 outcome. Prophylactic intervention of additional high-risk populations, according to their genetic profile, will be a key achievement for the fight against influenza viruses.
Similar content being viewed by others
Introduction
Influenza A viruses (single-stranded, negative-sense RNA viruses) are a major determinant of acute respiratory infections and an important cause of death worldwide. They are responsible not only for the recurrent epidemics but also for occasional pandemics due to major changes in antigenicity (as a result of mixing the segmented genome of two parent viruses of different origin, also called “antigenic shift”) leading up to half a million of people to death every year [1]. During the 2009 influenza pandemic, due to the emergence of a new influenza A virus subtype H1N1 [A(H1N1)pdm09] [2], Greece experienced two waves of transmission, where 294 patients received intensive care treatment and 149 patients died [3, 4]. In post-pandemic season 2010–11, pandemic influenza A(H1N1)pdm09 was by far the most commonly detected virus (97,5% in Greece) resulting to 368 admissions to intensive-care units (ICUs) and 180 deaths in Greece [5, 6].
Although seasonal influenza virus infection is in vast majority mild and limited in the upper respiratory tract, in a small subset of patients it can cause primary viral pneumonia as a complication and a minority may even evolve into acute respiratory distress syndrome (ARDS) and lead to death [7, 8]. Disease severity is usually associated with a plethora of host factors such as age (> 65), pregnancy, obesity, and underlying medical conditions including pulmonary, cardiovascular, renal, or hepatic disease, chronic metabolic disorder (including diabetes mellitus) or immunosuppression [9]. Moreover, the risk of a secondary bacterial pneumonia commonly caused by Streptococcus pneumoniae, Staphylococcus aureus, and Haemophilus influenzae is a key complication of influenza virus infection particularly in the elderly [10]. Notably, in the recent pandemic of 2009, 65% (80% of the respiratory and cardiovascular deaths) of influenza A(H1N1)pdm09 deaths worldwide were in individuals aged 18–64 years [11], whereas people over 65 years of age tended to have milder symptoms probably due to cross-protective immunity induced by previous H1N1 influenza infection or vaccination [12].
To understand the pathogenicity of influenza viruses, numerous parameters implicating both the host and the virus must be considered. These determinants are mainly the essential virulence of the virus, the acquired host immunity and the inheritable host factors whose importance in severity determination is only recently being underscored and granted major attention, especially after the emergence of the 2009 pandemic [13, 14]. The keystone that triggered studies on innate host factors was the observation of increased death rate among young adults and children without known risk factor mentioned before [15]. Using the genealogic database of families of Utah between 1904 and 2004, Albright et al. documented a significant association of influenza-associated death risk within families underlying the inherited predisposition of susceptibility to severe influenza infection [16]. In view of the contradictory results obtained by a second epidemiological study in Iceland with substantially smaller sample size [17], further investigation is needed to reveal the role of host genetic factors in the severity of influenza infections.
Recent technological advances have promoted the identification of host genetic factors involved in the life cycle of several viruses, such as HIV-1, HCV, Dengue virus, West Nile virus, CMV, RSV and HPV [18,19,20]. Similarly, a number of recent studies, using different strategies (genome-wide screens, proteomics or transcriptomics approaches, etc.) both in humans and in animal models, aim to reproduce and validate candidate genes retrieved mainly from genetic association studies that predispose to severe influenza [21,22,23]. Genetic polymorphisms vary among different individuals in each population and usually involve the substitution of a single nucleotide (single nucleotide polymorphism, SNP). Although the estimated SNP frequency within the human genome was about one SNP per 1000 base pairs, which corresponds to 3 million SNPs in total [24, 25], the international HapMap project reported about 10 million SNPs with a minor allele frequency (MAF) of at least 5% [26]. The different SNPs may affect in diverse ways the produced protein, and thus the phenotype, depending on their location: within a coding or non-coding sequence.
Several potential genetic polymorphisms predisposing for influenza severity have been recently described, including complement regulatory protein CD55; complement component 1, q subcomponent binding protein (C1QBP) gene; Fc fragment of immunoglobulin G, low-affinity IIA, receptor (FCGR2A) gene; interferon-inducible transmembrane protein 3 (IFITM3); tumor necrosis factor (TNF) gene; RPA interacting protein (RPAIN) gene; ST3 beta-galactosidase alpha − 2,3-sialyltransferase 1 (ST3GAL1) gene; lymphotoxin-alpha (LTA) gene and killer-cell immunoglobulin-like receptor (KIR) gene [22, 27,28,29,30,31].
CD55, or complement decay-accelerating factor (DAF), encodes a glycoprotein involved in the regulation of the complement cascade and participates in host cells protection from damage of pathogenic microorganisms [32]. Two further SNPs mapped in C1QBP (rs3786054) and FCGR2A (rs1801274) genes have been reported to associate with severe A influenza cases (ORs 3.13 and 2.68, respectively; P < 0.0001) in Mexico in 2009 [22]. C1QBP gene encodes a protein that binds to the globular heads of C1q molecules and can activate the classical pathway of complement [33], while FCGR2A encodes a member of a family of immunoglobulin Fc receptor genes found on the surface of phagocytic cells such as macrophages and neutrophils, and is involved in the process of phagocytosis and clearing of immune complexes [34].
In view of contradictory results observed in the current literature regarding the importance of host genetic polymorphisms in influenza severity, as well as their insufficient study, prompted us to investigate the association between SNPs of CD55 rs2564978, C1QBP rs3786054 and FCGR2A rs1801274 and severity of pandemic A(H1N1)pdm09 influenza in a European population. The polymorphisms selected are innate-immunity-related gene SNPs that were by previous genome-wide association studies (GWAS) initially proposed to be associated with severe A(H1N1)pdm09 infection, and their clinical significance needs to be validated. Thus, the aim of the present study has been to identify reliable host genetic risk factors associated with influenza A(H1N1)pdm09 outcome.
Methods
Patient samples
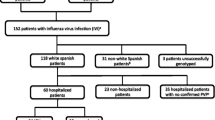
During the post-pandemic influenza period 2010–11, pharyngeal swabs or aspirate samples were sent to the National Influenza Center for Northern Greece for laboratory confirmation of influenza A(H1N1)pdm09 by one-step RT-PCR, according to a protocol developed by the Centre for Disease Control and Prevention [35]. In the present study, patients with confirmed A(H1N1)pdm09 influenza infection were recruited and classified as mild (control) or severe cases according to their clinical characteristics, such as, admission to the intensive care unit, requirement for oxygen supplementation and especially the presence or not of pneumonia according to the guidelines of the Infectious Diseases Society of America (IDSA) and the American Thoracic Society (ATS) [36] at baseline, as reported by individual clinicians. The samples were collected between January and February of the post-pandemic influenza period 2010–11 from 23 state hospitals and two Social Insurance Institutes (IKAs) from northern Greece. Moreover, to achieve uniformity in our study groups the following exclusion criteria were set: patients older than 60 years, other than Greek, or preexisting comorbidities in accordance to WHO risk factors. The demographic and clinical features of the study participants are shown in Table 1. The ethical approval of this study has been provided by the “Bioethics Committee of the Medical School of the Aristotle University of Thessaloniki” (protocol approval number 432).
Genotyping
Genomic DNA was extracted from patient samples using pharyngeal swabs or aspirate samples by the use of the QIAamp DNA Mini Kit (Qiagen, Germany). In brief, 1 ml of the sample was centrifuged at maximum speed (14,000 rpm) for 15 min and 200 µl of the precipitate was further processed following the manufacturer’s instructions. The concentration and the purity of the isolated DNAs were evaluated spectrophotometrically by the absorbance ratios at A260/A280 and A260/A230, respectively, using a Nanodrop ND-1000 (Thermo Fisher Scientific, Wilmington, USA). Extracted DNA samples were stored at − 20 °C until processing for genotyping analysis.
Allelic discrimination of three polymorphisms, CD55 rs2564978, C1QBP rs3786054 and FCGR2A rs1801274 was performed by real-time PCR using TaqMan technology on Applied Biosystems 7500 Fast Real-Time PCR system (Applied Biosystems Inc., CA, USA). Primers and probes were obtained from Applied Biosystems and available information to the SNPs is presented in Table 2. Genotyping of each sample was performed using the TaqMan Genotyping Master Mix in optically clear 96-well plates, in a final volume of 25 µl, containing 20 ng DNA. The cycling conditions were according to the manufacturer’s instructions.
Statistical analysis
Genotype and allele frequencies for the three SNPs were calculated by direct counting. Statistical analysis of the data was performed with SPSS 23.0 software (Chicago, Illinois, USA) using Fisher exact test. All SNPs were tested for Hardy–Weinberg equilibrium (HWE) using Exact test by PLINK software both in mild and severe cases. To investigate all genetic models of association, odds ratios (ORs) and their corresponding 95% confidence intervals (95% CI) were calculated according to dominant (heterozygotes plus minor allele homozygotes vs major allele homozygotes), co-dominant (heterozygotes vs minor allele homozygotes plus major allele homozygotes) and recessive (minor allele homozygotes vs major allele homozygotes plus heterozygotes) model using logistic regression analysis in SPSS 23.0. Additionally, the ORs and their corresponding 95% CIs under the homozygous, heterozygous and allelic models were calculated with logistic regression model using the glm function in R. Moreover, Tukey’s Honest Significant Difference (HSD) for post-hoc test was used to fix type I errors through the glht function of multcomp package [37]. P value < 0.05 was considered statistically significant for all analyses.
Results
Clinical and demographic features
Of the 810 positive for A(H1N1)pdm09 influenza samples in period 2010–11, 110 were selected according to their clinical and demographic features and classified as mild (control = 51) or severe cases (n = 59) that had radiographic signs of pneumonia. The average days between the onset of the symptoms and the presentation to the healthcare unit (sample collection) was 4 days. The design of the study and the criteria used for sample selection are described under “Methods” in section “Patient samples”. Male sex predominated in both groups (60.8% and 59.3%, respectively), while the mean age of the groups was 37.3 and 36.7 years of age, respectively (Table 1). Of interest, a very low vaccination rate (6.4% of all patients) was noticed in both groups and especially in severe cases, where only 3.4% were vaccinated for seasonal or pandemic influenza. As expected, the majority of fatal cases (85%) in the study had severe clinical signs of influenza infection and differed significantly from the control cases (P = 0.005). None of the study participants had any reported comorbidities or other influenza risk factors such as older age (> 65), pregnancy or obesity.
Distribution of the genotype and allele frequencies
Genotype and allele frequencies of CD55 (rs2564978), C1QBP (rs3786054) and FCGR2A (rs1801274) genes are presented in Table 3. Genotype distributions of all three SNPs were consistent with Hardy–Weinberg equilibrium in studied groups (P > 0.05) and their minor allelic frequencies (MAFs) were in accordance to those reported for European populations in the 1000 Genomes project data source (http://www.1000genomes.org). As shown in Table 3, there was no significant difference in genotype or allele frequencies of any polymorphism studied between mild and severe cases of influenza. On the contrary, when the distribution of genotype and allele frequencies in association with the mortality was investigated, there was a significant representation of the recessive genotype TT of CD55 rs2564978 (fatality rate 15.8%, P = 0.030) in the fatal cases against the non-fatal cases (2.2%). In addition, the percentage of the minor allele T of CD55 rs2564978 was also significantly increased in fatal cases (36.8%, P = 0.035) as compared to the non-fatal cases (20.3%) (Table 3). Similarly, patients bearing the recessive genotype AA (fatal cases 10.5% versus non-fatal cases 3.3%) as well as the heterozygous AG genotype (fatal cases 57.9% versus non-fatal cases 37.4%) of C1QBP rs3786054 were significantly increased (P = 0.042) in the fatal cases affecting also the frequency of the minor allele A (fatal cases 39.5% versus non-fatal cases 22%, P = 0.038) (Table 3). Regarding the FCGR2A rs1801274 polymorphism, although the frequency of the minor allele homozygotes GG increased, there was no genetic variation between the two groups analyzed (Table 3).
No significant differences in risk association of the three SNPs investigated were observed when patients with mild and severe influenza symptoms were compared under the dominant, co-dominant and recessive genetic models as shown in Table 4. Moreover, there were no differences observed even when the dominant genotype of each SNP was compared with its corresponding recessive genotype. Similarly, allelic odds ratios of CD55 rs2564978, C1QBP rs3786054 and FCGR2A rs1801274 revealed no differences in risk associated with influenza severity. The same analysis was carried out to uncover the genetic association between the three SNPs studied and the mortality risk. Indeed, minor allele homozygote patients of the CD55 rs2564978 presented 8.3 times increased risk (P = 0.026, OR 8.344, 95% CI 1.290–53.957) in fatal outcome than the major allele homozygotes plus heterozygotes under the recessive genetic model. The risk was further increased to 10.5-fold (P = 0.043, OR 10.500, 95% CI 1.062–103.788) when comparing the difference in odds between minor allele homozygotes vs major allele homozygotes. Additionally, under the allelic model of comparison, the patients carrying the T allele presented about 2.3 times increased risk (P = 0.031, OR 0.437, 95% CI 0.206–0.927) in fatal outcome compared to the patients with the C allele. Regarding C1QBP rs3786054, when comparing the major allele homozygotes GG with the minor allele homozygotes AA and heterozygotes AG under the dominant genetic model, the latter were found significantly associated with increased risk of mortality by threefold (P = 0.032, OR 3.162, 95% CI 1.102–9.072). This finding was further supported by the significant increase in odds of dying if harboring the minor allele A than the major allele G (P = 0.026, OR 0.432, 95% CI 0.206–0.904). In contrast, there was no significant difference in odds regarding the FCGR2A rs1801274 polymorphism under any genetic model examined. The results of association analysis under the seven different genetic models are summarized in Table 4.
Discussion
The emergence of the recent influenza pandemic in 2009 led more than 18,000 people (mostly young adults) to death worldwide [38]. In Greece, during the pandemic and the post-pandemic season, 329 people died, with the highest mortality rate being between 46 and 64 years of age [4, 5]. Indeed, in our study, the mean age of fatal cases was 48.21 years, while the mean ages of mild and severe cases with fatal outcome were 52.67 and 47.37 years, respectively. Another important issue to be highlighted is the very low vaccination rate in general population, although the influenza vaccination was offered for free in Greece. The vaccination coverage of our study population, which does not include high-risk groups, was 6.4%, a relatively low rate, but in accordance with the general population which was 5.9% that year [5]. Scientific and national health policies need to be applied, since low vaccination coverage may lead to epidemics, as it was observed in the recent measles epidemic in Europe.
The increased mortality rate in young adults triggered the present study to reveal the potential role of host genetics in the severity of illness as demonstrated by the presence of pneumonia or the fatal outcome. Towards that goal we decided to investigate the association between the innate-immunity-related gene SNPs of CD55 rs2564978, C1QBP rs3786054 and FCGR2A rs1801274 and the severity of pandemic A(H1N1)pdm09 influenza in a European population. It is interesting to note that amongst these three SNPs only in the first two of them we were able to obtain statistically significant data, as discussed below.
CD55 is a membrane-associated protein that regulates complement activation by interfering with C3/C5 convertases both in classical and alternative pathway, and naturally protects host cells from damage of pathogens. The TT genotype of rs2564978, that results in reduced promoter activity [29], is relatively rare in Europe (7%) according to the 1000 Genomes project data source and was found in only 4.5% of our overall patients (Table 3). However, there was a significant overrepresentation of the TT genotype in fatal cases that reached 15.8% (P = 0.030), and only 2.2% in survivors. Similar observation was seen by comparing the minor allele T distribution, since it was 36.8% in fatal cases vs 20.3% in non-fatal cases of our patient group. The resulted reduced expression of CD55 in patients carrying the rare TT genotype may lead to more robust complement activation during infection, resulting in worse outcomes secondary to enhanced inflammation [23]. Similarly, the death risk association was found significantly increased by more than eightfold in fatal cases (P = 0.026, OR 8.344, 95% CI 1.290–53.957), a result that is rather overestimated because of the small sample size in our study. Consistent with our findings, a recent study in Spain indirectly associated rs2564978 with influenza severity (P = 0.0064, OR 7.11, 95% CI 1.400–36.00) [39]. These findings are in accordance with those of Zhou and colleagues [29] that first described the significant association of the TT genotype with severe influenza infection (P = 0.011, OR 1.75, 95% CI 1.13–2.70) in Chinese patients based on a small-scale genome-wide association study (GWAS). Recently, a second study from China did not find any significant association between rs2564978 and the primary outcome in cases of avian (H7N9) or pandemic influenza, but TT genotype was linked to severity [40]. The fact that the TT genotype in East Asian populations is much more prevalent than in the rest of the world (35% vs 7%) according to the 1000 Genomes project data source, underlies the importance of this polymorphism in influenza severity and further attention is needed. Our study is the first presenting a positive correlation between fatal outcome and the TT genotype and this finding needs to be confirmed in a bigger cohort.
Apart from CD55, the study also focused on the association of host genetic polymorphisms with influenza severity of another protein related to complement cascade, since its role in viral defense is well established. The C1QBP gene encodes for a protein that binds to C1q molecule, a component known as an important regulator of antiviral antibody effector mechanisms of the classical pathway of complement activation, and thus inhibiting C1 [41]. C1QBP rs3786054 was one of the four SNPs (together with FCGR2A rs1801274, an unknown gene rs9856661 and RPAIN rs8070740) identified as significant risk factors on the severity of influenza infection in a case–control genetic association study in Mexican population. A threefold increase in risk of severe disease (P < 0.0001, OR 3.13, 95% CI 1.89–5.17) was found for patients homozygous for the minor allele [22]. Although we did not find any significant association of the rare AA genotype with severity in any genetic model studied, we found a threefold increased risk of death in dominant genetic model for the major G-allele (P = 0.032, OR 3.162, 95% CI 1.102–9.072). Moreover, the frequency of the rare AA genotype was significantly increased from 4.6% in all patients to 10.5% in patients who died. Additionally, the allele distribution of the minor allele A was also significantly (P = 0.038) increased in fatal cases (39.5%) as compared to non-fatal cases (22%). Unlikely, the association of rs3786054 with influenza severity could not be replicated in a recent GWAS [39] and thus in view of the contradictory results observed in different disease association studies, further research is required to clarify the role of this genetic polymorphism.
Another polymorphism, namely the rs1801274 on FCGR2A gene, was reported as candidate risk association factor in a study from Mexico [22]. The nonsynonymous A/G polymorphism rs1801274 causes an amino acid substitution at position 131 (His131Arg), which is known to affect the affinity of the FCGR2A for different subtypes of IgG [42]. The effector cells of AA homozygous individuals bind stronger to IgG2 resulting to more effective antigen clearance [43]. The enhanced host immune response might be the cause of severe disease outcome of A(H1N1)pdm09 influenza infection, taking into account that protein encoded by FCGR2A participates in the clearing of immune complexes [22, 23]. Zuniga and colleagues reported a significant increase of minor allele frequency in severe cases compared with control group (36% and 13%, respectively) as well as increased risk of association (P < 0.0001, OR 2.68, 95% CI 1.69–4.25) with severity. However, these results do not concur with our findings regarding either the presence of pneumonia or the risk of mortality where no association was established under any genetic model tested. In accordance with our findings, a recent study from Brazil failed to reveal any differences in allele or genotype frequencies of the rs1801274 polymorphism and its association with influenza disease severity or death (P > 0.05) [44]. Moreover, the association of rs1801274 with influenza severity was not even indirectly replicated in a European GWAS where more than half a million SNPs were investigated [39]. Probably the false discovery rate of 36% for rs1801274 in the study of Zuniga and colleagues, [22] because of the small size of the study, misled them to positive results. The main drawbacks of current studies is either the small sample size and/or the inability of generating compared groups for age, sex, ethnicity without confounding underlying medical conditions that question the importance of the findings and the overall polymorphism implication.
In this study, samples were collected during the short outbreak of A(H1N1)pdm09 influenza in the post pandemic period 2010–11. To eliminate ethnicity stratification that is an important confounding factor in genetic association studies, only Greek ethnicity patients were enrolled. Further, our study is restricted to patients without previous known influenza comorbidities, and cases were diagnosed with pneumonia. Such study design led to the inclusion of only 110 cases for further genotyping out of 810 cases examined. Our results show that two independent complement-related SNPs, namely the rare TT genotype of CD55 as well as the rare AA genotype of C1QBP are associated with increased death risk, but not with the disease severity based on the stratification sample criteria applied. No significant differences were observed for FCGR2A genotypes. Genotype distributions of all three SNPs investigated were consistent with Hardy–Weinberg equilibrium and their MAFs were in accordance to those reported for European populations in the 1000 Genomes project data source. Since all patients in our study had no other risk factors, our findings could lead to the identification of population groups at genetic risk. Additional genetic association studies could support our findings to the identification of new at-risk populations to whom the implementation of vaccination will help prevent influenza infection and its implications. Furthermore, their early recognition will reduce the financial burden due to less hospitalization.
References
WHO (2014) World health organization: influenza (seasonal). http://www.who.int/mediacentre/factsheets/fs211/en/. Accessed Sep 2017
Neumann G, Noda T, Kawaoka Y (2009) Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459(7249):931–939. https://doi.org/10.1038/nature08157
CDC (2010) Deaths and hospitalizations related to 2009 pandemic influenza A (H1N1)—Greece, May 2009–February 2010. MMWR Morb Mortal Wkly Rep 59(22):682–686
Athanasiou M, Lytras T, Spala G, Triantafyllou E, Gkolfinopoulou K, Theocharopoulos G, Patrinos S, Danis K, Detsis M, Tsiodras S, Bonovas S, Panagiotopoulos T (2010) Fatal cases associated with pandemic influenza A (H1N1) reported in Greece. PLoS Curr 2:RRN1194. https://doi.org/10.1371/currents.RRN1194
Athanasiou M, Baka A, Andreopoulou A, Spala G, Karageorgou K, Kostopoulos L, Patrinos S, Sideroglou T, Triantafyllou E, Mentis A, Malisiovas N, Lytras T, Tsiodras S, Panagiotopoulos T, Bonovas S (2011) Influenza surveillance during the post-pandemic influenza 2010/11 season in Greece, 04 October 2010–22 May 2011. Euro Surveill. https://doi.org/10.2807/ese.16.44.20004-en
Melidou A, Exindari M, Gioula G, Malisiovas N (2013) Severity of the two post-pandemic influenza seasons 2010–11 and 2011–12 in Northern Greece. Hippokratia 17(2):150–152
Hayward AC, Fragaszy EB, Bermingham A, Wang L, Copas A, Edmunds WJ, Ferguson N, Goonetilleke N, Harvey G, Kovar J, Lim MS, McMichael A, Millett ER, Nguyen-Van-Tam JS, Nazareth I, Pebody R, Tabassum F, Watson JM, Wurie FB, Johnson AM, Zambon M, Flu Watch G (2014) Comparative community burden and severity of seasonal and pandemic influenza: results of the flu watch cohort study. Lancet Respir Med 2(6):445–454. https://doi.org/10.1016/S2213-2600(14)70034-7
Cheng VC, To KK, Tse H, Hung IF, Yuen KY (2012) Two years after pandemic influenza A/2009/H1N1: what have we learned? Clin Microbiol Rev 25(2):223–263. https://doi.org/10.1128/CMR.05012-11
CDC (2017) People at high risk of developing flu-related complications. Centers for disease control and prevention. https://www.cdc.gov/flu/about/disease/high_risk.htm. Accessed Sep 2017
van Zyl G (2006) Laboratory Findings. In: Kamps B, Hoffmann C, Preiser W (eds) Influenza report 2006. Flying Publisher, Paris, pp 150–159
Dawood FS, Iuliano AD, Reed C, Meltzer MI, Shay DK, Cheng PY, Bandaranayake D, Breiman RF, Brooks WA, Buchy P, Feikin DR, Fowler KB, Gordon A, Hien NT, Horby P, Huang QS, Katz MA, Krishnan A, Lal R, Montgomery JM, Molbak K, Pebody R, Presanis AM, Razuri H, Steens A, Tinoco YO, Wallinga J, Yu H, Vong S, Bresee J, Widdowson MA (2012) Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis 12(9):687–695. https://doi.org/10.1016/S1473-3099(12)70121-4
Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, Brammer TL, Cox NJ, Tumpey TM, Katz JM (2009) Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med 361(20):1945–1952. https://doi.org/10.1056/NEJMoa0906453
Horby P, Nguyen NY, Dunstan SJ, Baillie JK (2013) An updated systematic review of the role of host genetics in susceptibility to influenza. Influenza Other Respir Viruses 7 Suppl 2:37–41. https://doi.org/10.1111/irv.12079
Horby P, Nguyen NY, Dunstan SJ, Baillie JK (2012) The role of host genetics in susceptibility to influenza: a systematic review. PLoS One 7(3):e33180. https://doi.org/10.1371/journal.pone.0033180
Ribeiro AF, Pellini AC, Kitagawa BY, Marques D, Madalosso G, de Cassia Nogueira Figueira G, Fred J, Albernaz RK, Carvalhanas TR, Zanetta DM (2015) Risk factors for death from Influenza A(H1N1)pdm09, state of Sao Paulo, Brazil, 2009. PLoS One 10(3):e0118772. https://doi.org/10.1371/journal.pone.0118772
Albright FS, Orlando P, Pavia AT, Jackson GG, Cannon Albright LA (2008) Evidence for a heritable predisposition to death due to influenza. J Infect Dis 197(1):18–24. https://doi.org/10.1086/524064
Gottfredsson M, Halldorsson BV, Jonsson S, Kristjansson M, Kristjansson K, Kristinsson KG, Love A, Blondal T, Viboud C, Thorvaldsson S, Helgason A, Gulcher JR, Stefansson K, Jonsdottir I (2008) Lessons from the past: familial aggregation analysis of fatal pandemic influenza (Spanish flu) in Iceland in 1918. Proc Natl Acad Sci USA 105(4):1303–1308. https://doi.org/10.1073/pnas.0707659105
Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ (2009) A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci USA 106(38):16410–16415. https://doi.org/10.1073/pnas.0907439106
Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139(7):1243–1254. https://doi.org/10.1016/j.cell.2009.12.017
König R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK (2010) Human host factors required for influenza virus replication. Nature 463(7282):813–817. https://doi.org/10.1038/nature08699
To KK, Zhou J, Chan JF, Yuen KY (2015) Host genes and influenza pathogenesis in humans: an emerging paradigm. Curr Opin Virol 14:7–15. https://doi.org/10.1016/j.coviro.2015.04.010
Zuniga J, Buendia-Roldan I, Zhao Y, Jimenez L, Torres D, Romo J, Ramirez G, Cruz A, Vargas-Alarcon G, Sheu CC, Chen F, Su L, Tager AM, Pardo A, Selman M, Christiani DC (2012) Genetic variants associated with severe pneumonia in A/H1N1 influenza infection. Eur Respir J 39(3):604–610. https://doi.org/10.1183/09031936.00020611
Lin TY, Brass AL (2013) Host genetic determinants of influenza pathogenicity. Curr Opin Virol 3(5):531–536. https://doi.org/10.1016/j.coviro.2013.07.005
Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, Hunt SE, Cole CG, Coggill PC, Rice CM, Ning Z, Rogers J, Bentley DR, Kwok PY, Mardis ER, Yeh RT, Schultz B, Cook L, Davenport R, Dante M, Fulton L, Hillier L, Waterston RH, McPherson JD, Gilman B, Schaffner S, Van Etten WJ, Reich D, Higgins J, Daly MJ, Blumenstiel B, Baldwin J, Stange-Thomann N, Zody MC, Linton L, Lander ES, Altshuler D, International SNPMWG (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409(6822):928–933. https://doi.org/10.1038/35057149
Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, Kruglyak L, Stein L, Hsie L, Topaloglou T, Hubbell E, Robinson E, Mittmann M, Morris MS, Shen N, Kilburn D, Rioux J, Nusbaum C, Rozen S, Hudson TJ, Lipshutz R, Chee M, Lander ES (1998) Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280(5366):1077–1082
International HapMap C, Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, Dermitzakis E, Bonnen PE, Altshuler DM, Gibbs RA, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Yu F, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Gibbs RA, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Dermitzakis E, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Bonnen PE, Gibbs RA, Gonzaga-Jauregui C, Keinan A, Price AL, Yu F, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Schaffner SF, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Pollack S, Price AL, Schaffner SF, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE (2010) Integrating common and rare genetic variation in diverse human populations. Nature 467(7311):52–58. https://doi.org/10.1038/nature09298
Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, Wise HM, Kane L, Goulding D, Digard P, Anttila V, Baillie JK, Walsh TS, Hume DA, Palotie A, Xue Y, Colonna V, Tyler-Smith C, Dunning J, Gordon SB, Smyth RL, Openshaw PJ, Dougan G, Brass AL, Kellam P (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484(7395):519–523. https://doi.org/10.1038/nature10921
Antonopoulou A, Baziaka F, Tsaganos T, Raftogiannis M, Koutoukas P, Spyridaki A, Mouktaroudi M, Kotsaki A, Savva A, Georgitsi M, Giamarellos-Bourboulis EJ (2012) Role of tumor necrosis factor gene single nucleotide polymorphisms in the natural course of 2009 influenza A H1N1 virus infection. Int J Infect Dis 16(3):e204–e208. https://doi.org/10.1016/j.ijid.2011.11.012
Zhou J, To KK, Dong H, Cheng ZS, Lau CC, Poon VK, Fan YH, Song YQ, Tse H, Chan KH, Zheng BJ, Zhao GP, Yuen KY (2012) A functional variation in CD55 increases the severity of 2009 pandemic H1N1 influenza A virus infection. J Infect Dis 206(4):495–503. https://doi.org/10.1093/infdis/jis378
Maestri A, Sortica VA, Tovo-Rodrigues L, Santos MC, Barbagelata L, Moraes MR, Alencar de Mello W, Gusmao L, Sousa RC, Emanuel Batista Dos Santos S (2015) Siaalpha2-3Galbeta1- receptor genetic variants are associated with influenza A(H1N1)pdm09 severity. PLoS One 10(10):e0139681. https://doi.org/10.1371/journal.pone.0139681
Morales-Garcia G, Falfan-Valencia R, Garcia-Ramirez RA, Camarena A, Ramirez-Venegas A, Castillejos-Lopez M, Perez-Rodriguez M, Gonzalez-Bonilla C, Grajales-Muniz C, Borja-Aburto V, Mejia-Arangure JM (2012) Pandemic influenza A/H1N1 virus infection and TNF, LTA, IL1B, IL6, IL8, and CCL polymorphisms in Mexican population: a case–control study. BMC Infect Dis 12:299. https://doi.org/10.1186/1471-2334-12-299
Zhang Y, Zhang Z, Cao L, Lin J, Yang Z, Zhang X (2017) A common CD55 rs2564978 variant is associated with the susceptibility of non-small cell lung cancer. Oncotarget 8(4):6216–6221. https://doi.org/10.18632/oncotarget.14053
Kishore U, Reid KB (2000) C1q: structure, function, and receptors. Immunopharmacology 49(1–2):159–170
Bermejo-Martin JF, Martin-Loeches I, Rello J, Anton A, Almansa R, Xu L, Lopez-Campos G, Pumarola T, Ran L, Ramirez P, Banner D, Ng DC, Socias L, Loza A, Andaluz D, Maravi E, Gomez-Sanchez MJ, Gordon M, Gallegos MC, Fernandez V, Aldunate S, Leon C, Merino P, Blanco J, Martin-Sanchez F, Rico L, Varillas D, Iglesias V, Marcos MA, Gandia F, Bobillo F, Nogueira B, Rojo S, Resino S, Castro C, Ortiz de Lejarazu R, Kelvin D (2010) Host adaptive immunity deficiency in severe pandemic influenza. Crit Care 14(5):R167. https://doi.org/10.1186/cc9259
CDC (2009) CDC protocol of realtime RTPCR for swine influenza A(H1N1). World health organisation. http://www.who.int/csr/resources/publications/swineflu/CDCrealtimeRTPCRprotocol_20090428.pdf. Accessed Sep 2017
Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, Dowell SF, File TM Jr, Musher DM, Niederman MS, Torres A, Whitney CG,, American Thoracic S (2007) Infectious diseases society of A. infectious diseases society of America/American thoracic society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis 44(Suppl 2):S27–S72. https://doi.org/10.1086/511159
Bretz F, Hothorn T, Westfall P (2010) Multiple comparisons using R. Chapman and Hall/CRC, Boca Raton
WHO (2009) Pandemic (H1N1) 2009—update 109. WHO global alert and response [online]. http://www.who.int/csr/don/2010_07_16/en/index.html Accessed Sep 2017
Garcia-Etxebarria K, Bracho MA, Galan JC, Pumarola T, Castilla J, Ortiz de Lejarazu R, Rodriguez-Dominguez M, Quintela I, Bonet N, Garcia-Garcera M, Dominguez A, Gonzalez-Candelas F, Calafell F, Cases C, Controls in Pandemic Influenza Working G (2015) No major host genetic risk factor contributed to A(H1N1)2009 influenza severity. PLoS One 10(9):e0135983. https://doi.org/10.1371/journal.pone.0135983
Lee N, Cao B, Ke C, Lu H, Hu Y, Tam CHT, Ma RCW, Guan D, Zhu Z, Li H, Lin M, Wong RYK, Yung IMH, Hung TN, Kwok K, Horby P, Hui DSC, Chan MCW, Chan PKS (2017) IFITM3, TLR3, and CD55 gene SNPs and cumulative genetic risks for severe outcomes in Chinese patients with H7N9/H1N1pdm09 influenza. J Infect Dis 216(1):97–104. https://doi.org/10.1093/infdis/jix235
Stoermer KA, Morrison TE (2011) Complement and viral pathogenesis. Virology 411(2):362–373. https://doi.org/10.1016/j.virol.2010.12.045
Clark MR, Stuart SG, Kimberly RP, Ory PA, Goldstein IM (1991) A single amino acid distinguishes the high-responder from the low-responder form of Fc receptor II on human monocytes. Eur J Immunol 21(8):1911–1916. https://doi.org/10.1002/eji.1830210820
van Sorge NM, van der Pol WL, van de Winkel JG (2003) FcgammaR polymorphisms: Implications for function, disease susceptibility and immunotherapy. Tissue Antigens 61(3):189–202
Maestri A, Sortica VA, Ferreira DL, de Almeida Ferreira J, Amador MA, de Mello WA, Santos SE, Sousa RC (2016) The His131Arg substitution in the FCGR2A gene (rs1801274) is not associated with the severity of influenza A(H1N1)pdm09 infection. BMC Res Notes 9:296. https://doi.org/10.1186/s13104-016-2096-1
Funding
This work received no specific grant from any funding agency.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures included in this work were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. To this end, the ethical approval of this study has been provided by the “Bioethics Committee of the Medical School of the Aristotle University of Thessaloniki” (protocol approval number 432).
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Edited by: S. Becker.
Rights and permissions
About this article
Cite this article
Chatzopoulou, F., Gioula, G., Kioumis, I. et al. Identification of complement-related host genetic risk factors associated with influenza A(H1N1)pdm09 outcome: challenges ahead. Med Microbiol Immunol 208, 631–640 (2019). https://doi.org/10.1007/s00430-018-0567-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00430-018-0567-9