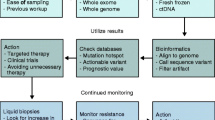
Abstract
Precision and personalized therapeutics have witnessed significant advancements in technology, revolutionizing the capabilities of laboratories to generate vast amounts of genetic data. Coupled with computational resources for analysis and interpretation, and integrated with various other types of data, including genomic data, electronic medical health (EMH) data, and clinical knowledge, these advancements support optimized health decisions. Among these technologies, next-generation sequencing (NGS) stands out as a transformative tool in the field of cancer treatment, playing a crucial role in precision oncology. NGS-based workflows are employed across a range of applications, including gene panels, exome sequencing, and whole-genome sequencing, supporting comprehensive analysis of the entire cancer genome, including mutations, copy number variations, gene expression profiles, and epigenetic modifications. By utilizing the power of NGS, these workflows contribute to enhancing our understanding of disease mechanisms, diagnosis confirmation, identifying therapeutic targets, and guiding personalized treatment decisions. This manuscript explores the diverse applications of NGS in cancer treatment, highlighting its significance in guiding diagnosis and treatment decisions, identifying therapeutic targets, monitoring disease progression, and improving patient outcomes.



Similar content being viewed by others
References
Singal G et al (2019) Association of patient characteristics and tumor genomics with clinical outcomes among patients with non-small cell lung cancer using a clinicogenomic database. JAMA 321(14):1391–1399
Levy SE, Myers RM (2016) Advancements in next-generation sequencing. Annu Rev Genomics Hum Genet 17:95–115
Satam H et al (2023) Next-generation sequencing technology: current trends and advancements. Biology (Basel) 12:7
Goodwin S, McPherson JD, McCombie WR (2016) Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 17(6):333–351
Rothberg JM et al (2011) An integrated semiconductor device enabling non-optical genome sequencing. Nature 475(7356):348–352
Uhlen M, Quake SR (2023) Sequential sequencing by synthesis and the next-generation sequencing revolution. Trends Biotechnol 41(12):1565–1572
van Dijk EL, Naquin D, Gorrichon K, Jaszczyszyn Y, Ouazahrou R, Thermes C, Hernandez C (2023) Genomics in the long-read sequencing era. Trends Genet 39(9):649–671
Rhoads A, Au KF (2015) PacBio sequencing and its applications. Genom Proteomics Bioinforma 13(5):278–289
Eid J et al (2009) Real-time DNA sequencing from single polymerase molecules. Science 323(5910):133–138
Mikheyev AS, Tin MM (2014) A first look at the Oxford Nanopore MinION sequencer. Mol Ecol Resour 14(6):1097–1102
Quick J, Quinlan AR, Loman NJ (2014) A reference bacterial genome dataset generated on the MinION portable single-molecule nanopore sequencer. Gigascience 3:22
Chaisson MJ et al (2015) Resolving the complexity of the human genome using single-molecule sequencing. Nature 517(7536):608–611
Wenger AM et al (2019) Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol 37(10):1155–1162
Vollger MR et al (2020) Improved assembly and variant detection of a haploid human genome using single-molecule, high-fidelity long reads. Ann Hum Genet 84(2):125–140
Volden R et al (2018) Improving nanopore read accuracy with the R2C2 method enables the sequencing of highly multiplexed full-length single-cell cDNA. Proc Natl Acad Sci U S A 115(39):9726–9731
Lee I et al (2020) Simultaneous profiling of chromatin accessibility and methylation on human cell lines with nanopore sequencing. Nat Methods 17(12):1191–1199
Simpson JT et al (2017) Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods 14(4):407–410
Vaser R et al (2017) Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27(5):737–746
Bonenfant Q, Noe L, Touzet H (2023) Porechop_ABI: discovering unknown adapters in Oxford Nanopore Technology sequencing reads for downstream trimming. Bioinform Adv 3(1):vbac085
Salmela L, Rivals E (2014) LoRDEC: accurate and efficient long read error correction. Bioinformatics 30(24):3506–3514
Yang X et al (2013) V-Phaser 2: variant inference for viral populations. BMC Genomics 14:674
Boza V, Brejova B, Vinar T (2017) DeepNano: deep recurrent neural networks for base calling in MinION nanopore reads. PLoS ONE 12(6):e0178751
Rang FJ, Kloosterman WP, de Ridder J (2018) From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol 19(1):90
Wang X, Liotta L (2011) Clinical bioinformatics: a new emerging science. J Clin Bioinforma 1(1):1
Roy S et al (2018) Standards and guidelines for validating next-generation sequencing bioinformatics pipelines: a joint recommendation of the Association for Molecular Pathology and the College of American Pathologists. J Mol Diagn 20(1):4–27
Roy S et al (2016) Next-generation sequencing informatics: challenges and strategies for implementation in a clinical environment. Arch Pathol Lab Med 140(9):958–975
Thankaswamy-Kosalai S, Sen P, Nookaew I (2017) Evaluation and assessment of read-mapping by multiple next-generation sequencing aligners based on genome-wide characteristics. Genomics 109(3–4):186–191
Cibulskis K et al (2013) Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31(3):213–219
Koboldt DC et al (2012) VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22(3):568–576
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38(16):e164
den Dunnen JT et al (2016) HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat 37(6):564–569
Li MM et al (2017) Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 19(1):4–23
Robinson JT et al (2011) Integrative genomics viewer. Nat Biotechnol 29(1):24–26
Sayers EW, O’Sullivan C, Karsch-Mizrachi I (2022) Using GenBank and SRA methods. Mol Biol 2443:1–25
Kadri S et al (2022) Containers in bioinformatics: applications, practical considerations, and best practices in molecular pathology. J Mol Diagn 24(5):442–454
Conway JR, Warner JL, Rubinstein WS, Miller RS (2019) Next-generation sequencing and the clinical oncology workflow: data challenges, proposed solutions, and a call to action. JCO Precis Oncol 3:PO.19.00232
Heeke AL, Pishvaian MJ, Lynce F, Xiu J, Brody JR, Chen WJ, Baker TM, Marshall JL, Isaacs C (2018) Prevalence of homologous recombination-related gene mutations across multiple cancer types. JCO Precis Oncol 2018
Pratt D, Sahm F, Aldape K (2021) DNA methylation profiling as a model for discovery and precision diagnostics in neuro-oncology. Neuro Oncol 23(23 5):S16–S29
Alexandrov LB et al (2020) The repertoire of mutational signatures in human cancer. Nature 578(7793):94–101
Rekhtman N et al (2020) SMARCA4-deficient thoracic sarcomatoid tumors represent primarily smoking-related undifferentiated carcinomas rather than primary thoracic sarcomas. J Thorac Oncol 15(2):231–247
Fumet JD et al (2020) Tumour mutational burden as a biomarker for immunotherapy: Current data and emerging concepts. Eur J Cancer 131:40–50
Rizvi NA et al (2015) Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348(6230):124–8
Sha D et al (2020) Tumor mutational burden as a predictive biomarker in solid tumors. Cancer Discov 10(12):1808–1825
Sholl LM et al (2020) The promises and challenges of tumor mutation burden as an immunotherapy biomarker: a perspective from the International Association for the Study of Lung Cancer Pathology Committee. J Thorac Oncol 15(9):1409–1424
Jardim DL et al (2021) The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell 39(2):154–173
Samstein RM et al (2019) Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 51(2):202–206
Frampton GM et al (2013) Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31(11):1023–1031
Chaudhary R et al (2018) A scalable solution for tumor mutational burden from formalin-fixed, paraffin-embedded samples using the oncomine tumor mutation load assay. Transl Lung Cancer Res 7(6):616–630
Vanderwalde A et al (2018) Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med 7(3):746–756
Buchhalter I et al (2019) Size matters: dissecting key parameters for panel-based tumor mutational burden analysis. Int J Cancer 144(4):848–858
Snyder A et al (2014) Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371(23):2189–2199
Garon EB et al (2019) Five-year overall survival for patients with advanced non-small-cell lung cancer treated with pembrolizumab: results from the phase I KEYNOTE-001 Study. J Clin Oncol 37(28):2518–2527
McGrail DJ et al (2021) High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol 32(5):661–672
Portela A, Esteller M (2010) Epigenetic modifications and human disease. Nat Biotechnol 28(10):1057–1068
Song K, Xu H, Wang C (2022) The role of N6-methyladenosine methylation in the progression of endometrial cancer. Cancer Biother Radiopharm 37(9):737–749
Moghbeli M et al (2014) Role of hMLH1 and E-cadherin promoter methylation in gastric cancer progression. J Gastrointest Cancer 45(1):40–47
Fujimoto M et al (2005) Methylation adjacent to negatively regulating AP-1 site reactivates TrkA gene expression during cancer progression. Oncogene 24(32):5108–5118
Papanicolau-Sengos A, Aldape K (2022) DNA methylation profiling: an emerging paradigm for cancer diagnosis. Annu Rev Pathol 17:295–321
Koelsche C, von Deimling A (2022) Methylation classifiers: brain tumors, sarcomas, and what’s next. Genes Chromosomes Cancer 61(6):346–355
Asaoka Y, Ijichi H, Koike K (2015) PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 373(20):1979
Petruzzellis G et al (2019) Role of DNA methylation profile in diagnosing astroblastoma: a case report and literature review. Front Genet 10:391
Capper D et al (2018) DNA methylation-based classification of central nervous system tumours. Nature 555(7697):469–474
Sturm D et al (2016) New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164(5):1060–1072
Lehman NL et al (2019) Genomic analysis demonstrates that histologically-defined astroblastomas are molecularly heterogeneous and that tumors with MN1 rearrangement exhibit the most favorable prognosis. Acta Neuropathol Commun 7(1):42
Mackinnon AC Jr, Johnson CM, Robin A, Christiansen L, Hanbazazh M, Summey RM, Chandrashaker D, Harada S, Bradley WH (2023) Pathologic, immunologic, and clinical analysis of the microsatellite instability phenotype in endometrial carcinoma. Hum Pathol 139:80–90
Zhu L et al (2018) A novel and reliable method to detect microsatellite instability in colorectal cancer by next-generation sequencing. J Mol Diagn 20(2):225–231
Stelloo E et al (2017) Practical guidance for mismatch repair-deficiency testing in endometrial cancer. Ann Oncol 28(1):96–102
Wang Y et al (2017) Differences in microsatellite instability profiles between endometrioid and colorectal cancers: a potential cause for false-negative results? J Mol Diagn 19(1):57–64
Mackinnon AC Jr et al (2023) Pathologic, immunologic, and clinical analysis of the microsatellite instability phenotype in endometrial carcinoma. Hum Pathol 139:80–90
Le DT et al (2017) Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357(6349):409–413
Le DT et al (2015) PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372(26):2509–2520
Kahn RM et al (2019) Universal endometrial cancer tumor typing: how much has immunohistochemistry, microsatellite instability, and MLH1 methylation improved the diagnosis of Lynch syndrome across the population? Cancer 125(18):3172–3183
Nguyen L et al (2020) Pan-cancer landscape of homologous recombination deficiency. Nat Commun 11(1):5584
Maxwell KN et al (2017) BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat Commun 8(1):319
Poti A et al (2019) Correlation of homologous recombination deficiency induced mutational signatures with sensitivity to PARP inhibitors and cytotoxic agents. Genome Biol 20(1):240
Pacheco-Barcia V et al (2022) The homologous recombination deficiency scar in advanced cancer: agnostic targeting of damaged DNA repair. Cancers (Basel) 14:12
Telli ML et al (2016) Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res 22(15):3764–3773
Lawrence MS et al (2013) Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499(7457):214–218
Ding L et al (2012) Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481(7382):506–510
Liu Z et al (2022) Towards accurate and reliable resolution of structural variants for clinical diagnosis. Genome Biol 23(1):68
Robbe P et al (2018) Clinical whole-genome sequencing from routine formalin-fixed, paraffin-embedded specimens: pilot study for the 100,000 genomes project. Genet Med 20(10):1196–1205
Duncavage EJ et al (2021) Genome sequencing as an alternative to cytogenetic analysis in myeloid cancers. N Engl J Med 384(10):924–935
Dorney R et al (2023) Recent advances in cancer fusion transcript detection. Brief Bioinform 24:1
Heydt C et al (2021) Detection of gene fusions using targeted next-generation sequencing: a comparative evaluation. BMC Med Genomics 14(1):62
Kim T et al (2020) RNA sequencing as an alternative tool for detecting measurable residual disease in core-binding factor acute myeloid leukemia. Sci Rep 10(1):20119
Mandelker D, Ceyhan-Birsoy O (2020) Evolving significance of tumor-normal sequencing in cancer care. Trends Cancer 6(1):31–39
Dumbrava EI, Brusco L, Daniels M, Wathoo C, Shaw K, Lu K, Zheng X, Strong L, Litton J, Arun B, Eterovic AK, Routbort M, Patel K, Qi Y, Piha-Paul S, Subbiah V, Hong D, Rodon J, Kopetz S, Mendelsohn J, Mills GB, Chen K, Meric-Bernstam F (2019) Expanded analysis of secondary germline findings from matched tumor/normal sequencing identifies additional clinically significant mutations. JCO Precis Oncol 3:PO.18.00143
Mandelker D, Zhang L (2018) The emerging significance of secondary germline testing in cancer genomics. J Pathol 244(5):610–615
Sahajpal NS et al (2021) Optical genome mapping as a next-generation cytogenomic tool for detection of structural and copy number variations for prenatal genomic analyses. Genes (Basel) 12:3
Sahajpal NS et al (2023) Clinical utility of optical genome mapping and 523-gene next generation sequencing panel for comprehensive evaluation of myeloid cancers. Cancers (Basel) 15:12
Levy B et al (2023) Optical genome mapping in acute myeloid leukemia: a multicenter evaluation. Blood Adv 7(7):1297–1307
Neveling K et al (2021) Next-generation cytogenetics: comprehensive assessment of 52 hematological malignancy genomes by optical genome mapping. Am J Hum Genet 108(8):1423–1435
Incorvaia L, Russo A, Cinieri S (2022) The molecular tumor board: a tool for the governance of precision oncology in the real world. Tumori 108(4):288–290
Tamborero D et al (2022) The Molecular Tumor Board Portal supports clinical decisions and automated reporting for precision oncology. Nat Cancer 3(2):251–261
Cannon TL et al (2022) Patient attendance at molecular tumor board: a new means of shared decision making? Curr Probl Cancer 46(3):100860
Nierengarten MB (2023) MatchMiner open-source platform matches patients with cancer to precision medicine trials. Cancer 129(4):494
Author information
Authors and Affiliations
Contributions
The contribution of both authors was equal, reflecting a collaborative and balanced effort in the development of this work.
Corresponding authors
Ethics declarations
Ethical approval
We affirm that both authors have adhered to ethical standards throughout the creation of this manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mackinnon, A.C., Chandrashekar, D.S. & Suster, D.I. Molecular pathology as basis for timely cancer diagnosis and therapy. Virchows Arch 484, 155–168 (2024). https://doi.org/10.1007/s00428-023-03707-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-023-03707-2