Abstract
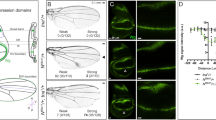
The tantalus (tan) gene encodes a protein that interacts specifically with the Polycomb/trithorax group protein Additional sex combs (ASX). Both loss-of-function and gain-of-function mutations in tan cause tissue-specific defects in the eyes, wing veins and bristles of adult flies. As these defects are also typical for components of the Notch (N) signalling pathway, we wished to determine if TAN interacts with this pathway. Through careful examination of ectopic tan phenotypes, we find that TAN specifically disrupts all three major processes associated with the N signalling pathway (boundary formation, lateral inhibition, and lineage decisions). Furthermore, ectopic tan expression abolishes expression of two N target genes, wingless (wg) and cut, at the dorsal-ventral boundary of the wing. An interaction between tan and N was also observed using a genetic assay that previously detected interactions between tan and Asx. The previously observed ability of TAN to move between the cytoplasm and nucleus, and to associate with DNA, provides a potential mechanism for TAN to respond to N signalling.


Similar content being viewed by others
References
Abdelilah-Seyfried S, Chan YM, Zeng C, Justice NJ, Younger-Shepherd S, Sharp LE, Barbel S, Meadows SA, Jan LY, Jan YN (2000) A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics 155:733–752
Bellen HJ, O’Kane CJ, Wilson C, Grossniklaus U, Pearson RK, Gehring WJ (1989) P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev 3:1288–1300
Bray S (1998) Notch signalling in Drosophila: three ways to use a pathway. Semin Cell Dev Biol 9:591–597
Carroll SB, Whyte JS (1989) The role of the hairy gene during Drosophila morphogenesis: stripes in imaginal discs. Genes Dev 3:905–916
de Celis (1998) Positioning and differentiation of veins in the Drosophila wing. Int J Dev Biol 42:335–343
de Celis JF, Tyler DM, de Celis J, Bray SJ (1998) Notch signalling mediates segmentation of the Drosophila leg. Development 125:4617–4626
Dietrich BH, Moore J, Kyba M, dosSantos G, McCloskey F, Milne TA, Brock HW, Krause HM (2001) Tantalus, a novel ASX-interacting protein with tissue-specific functions. Dev Biol 234:441–453
Kadesch T (2000) Notch signalling: a dance of proteins changing partners. Exp Cell Res 260:1–8
Klein T, Seugnet L, Haenlin M, Martinez-Arias A (2000) Two different activities of Suppressor of Hairless during wing development in Drosophila. Development 127:3553–3566
Lai EC (2004) Notch signaling: control of cell communication and cell fate. Development 131:965–973
Ligoxygakis P, Bray SJ, Apidianakis Y, Delidakis C (1999) Ectopic expression of individual E(spl) genes has differential effects on different cell fate decisions and underscores the biphasic requirement for Notch activity in wing margin establishment in Drosophila. Development 126:2205–2214
Mazaleyrat SL, Fostier M, Wilkin MB, Aslam H, Evans DA, Cornell M, Baron M (2003) Down-regulation of Notch target gene expression by suppressor of deltex. Dev Biol 255:363–372
Mishra A, Agrawal N, Banerjee S, Sardesai D, Dalal JS, Bhojwani J, Sinha P (2001) Spatial regulation of DELTA expression mediates NOTCH signalling for segmentation of Drosophila legs. Mech Dev 105:115–127
Orlando V (2003) Polycomb, epigenomes, and control of cell identity. Cell 112:599–606
Posakony JW (1994) Nature versus nurture: asymmetric cell divisions in Drosophila bristle development. Cell 76:415–418
Acknowledgements
We thank the Bloomington Stock Center and G. Boulianne for fly stocks, and G. Boulianne for comments on the manuscript. This work was supported by funds provided by the National Cancer Institute of Canada (to H.M.K.) and a University of Toronto Open Fellowship (to B.H.D.). B.H.D. would like to thank N. Dostatni for allowing part of this work to be performed in her laboratory.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by P. Simpson
Rights and permissions
About this article
Cite this article
Dietrich, B.H., Yang, P. & Krause, H.M. tantalus, a potential link between Notch signalling and chromatin-remodelling complexes. Dev Genes Evol 215, 255–260 (2005). https://doi.org/10.1007/s00427-005-0471-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-005-0471-3