Abstract
Main conclusion
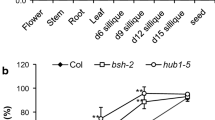
WOX family gene WOX2 is highly expressed during seed development, which functions redundantly with WOX1 and WOX4 to positively regulate seed germination.
Abstract
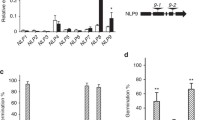
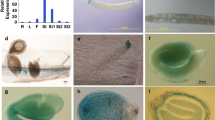
WOX (WUSCHEL-related homeobox) is a family of transcription factors in plants. They play essential roles in the regulation of plant growth and development, but their function in seed germination is not well understood. In this report, we show that WOX1, WOX2, and WOX4 are close homologues in Arabidopsis. WOX2 has a redundant function with WOX1 and WOX4, respectively, in seed germination. WOX2 is highly expressed during seed development, from the globular embryonic stage to mature dry seeds, and its expression is decreased after germination. Loss of function single mutant wox2, and double mutants wox1 wox2 and wox2 wox4-1 show decreased germination speed. WOX2 and WOX4 are essential for hypocotyl–radicle zone elongation during germination, potentially by promoting the expression of cell wall-related genes. We also found that WOX2 and WOX4 regulate germination through the gibberellin (GA) pathway. These results suggest that WOX2 and WOX4 integrate the GA pathway and downstream cell wall-related genes during germination.








Similar content being viewed by others
Data availability
All data in this study are provided in this manuscript and supplementary data files. It will be provided upon a reasonable request.
Abbreviations
- DAP:
-
Days after pollination
- GA:
-
Gibberellin
- GO:
-
Gene Ontology
- GUS:
-
β-Glucuronidase
- PAC:
-
Paclobutrazol
- WOX:
-
WUSCHEL-related homeobox
- XTH:
-
Xyloglucan endotransglucosylase/hydrolase
References
Bassel GW (2016) To grow or not to grow? Trends Plant Sci 21:498–505. https://doi.org/10.1016/j.tplants.2016.02.001
Bassel GW, Stamm P, Mosca G, Barbier de Reuille P, Gibbs DJ, Winter R, Janka A, Holdsworth MJ, Smith RS (2014) Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo. Proc Natl Acad Sci USA 111:8685–8690. https://doi.org/10.1073/pnas.1404616111
Becnel J, Natarajan M, Kipp A, Braam J (2006) Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol Biol 61:451–467. https://doi.org/10.1007/s11103-006-0021-z
Brackmann K, Qi J, Gebert M, Jouannet V, Schlamp T, Grunwald K, Wallner ES, Novikova DD, Levitsky VG, Agusti J, Sanchez P, Lohmann JU, Greb T (2018) Spatial specificity of auxin responses coordinates wood formation. Nat Commun 9:875. https://doi.org/10.1038/s41467-018-03256-2
Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T (2008) Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev Cell 14:867–876. https://doi.org/10.1016/j.devcel.2008.03.008
Burkart RC, Strotmann VI, Kirschner GK, Akinci A, Czempik L, Dolata A, Maizel A, Weidtkamp-Peters S, Stahl Y (2022) PLETHORA-WOX5 interaction and subnuclear localization control Arabidopsis root stem cell maintenance. EMBO Rep 23:e54105. https://doi.org/10.15252/embr.202154105
Cao XW, He ZS, Guo L, Liu XG (2015) Epigenetic mechanisms are critical for the regulation of WUSCHEL expression in floral meristems. Plant Physiol 168:1189–1196. https://doi.org/10.1104/pp.15.00230
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. https://doi.org/10.1046/j.1365-313x.1998.00343.x
Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6:850–861. https://doi.org/10.1038/nrm1746
Deveaux Y, Toffano-Nioche C, Claisse G, Thareau V, Morin H, Laufs P, Moreau H, Kreis M, Lecharny A (2008) Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol Biol 8:291. https://doi.org/10.1186/1471-2148-8-291
Dhar S, Kim J, Yoon EK, Jang S, Ko K, Lim J (2022) SHORT-ROOT controls cell elongation in the etiolated Arabidopsis hypocotyl. Mol Cells 45:243–256. https://doi.org/10.14348/molcells.2021.5008
Etchells JP, Provost CM, Mishra L, Turner SR (2013) WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development 140:2224–2234. https://doi.org/10.1242/dev.091314
Forzani C, Aichinger E, Sornay E, Willemsen V, Laux T, Dewitte W, Murray JA (2014) WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Curr Biol 24:1939–1944. https://doi.org/10.1016/j.cub.2014.07.019
Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Höfte H (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol 114:295–305. https://doi.org/10.1104/pp.114.1.295
Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6:609–618. https://doi.org/10.1111/j.1467-7652.2008.00346.x
Holdsworth MJ, Bentsink L, Soppe WJJ (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179:33–54. https://doi.org/10.1111/j.1469-8137.2008.02437.x
Hu X, Xu L (2016) Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol 172:2363–2373. https://doi.org/10.1104/pp.16.01067
Jha P, Ochatt SJ, Kumar V (2020) WUSCHEL: a master regulator in plant growth signaling. Plant Cell Rep 39:431–444. https://doi.org/10.1007/s00299-020-02511-5
Kong X, Lu S, Tian H, Ding Z (2015) WOX5 is shining in the root stem cell niche. Trends Plant Sci 20:601–603. https://doi.org/10.1016/j.tplants.2015.08.009
Kong D, Hao Y, Cui H (2016) The WUSCHEL related homeobox protein WOX7 regulates the sugar response of lateral root development in Arabidopsis thaliana. Mol Plant 9:261–270. https://doi.org/10.1016/j.molp.2015.11.006
Kong X, Li C, Zhang F, Yu Q, Gao S, Zhang M, Tian H, Zhang J, Yuan X, Ding Z (2018) Ethylene promotes cadmium-induced root growth inhibition through EIN3 controlled XTH33 and LSU1 expression in Arabidopsis. Plant Cell Environ 41:2449–2462. https://doi.org/10.1111/pce.13361
Kucukoglu M, Nilsson J, Zheng B, Chaabouni S, Nilsson O (2017) WUSCHEL-RELATED HOMEOBOX4 (WOX4)-like genes regulate cambial cell division activity and secondary growth in Populus trees. New Phytol 215:642–657. https://doi.org/10.1111/nph.14631
Liao JK, Deng B, Cai XY, Yang QX, Hu BP, Cong JJ, Zhang YX, Wang G, Xin GL, Li YT, Yang L, Zhang DZ, Zhang J, Liu BB (2022) Time-course transcriptome analysis reveals regulation of Arabidopsis seed dormancy by the transcription factors WOX11/12. J Exp Bot 74:1090–1106. https://doi.org/10.1093/jxb/erac457
Lie C, Kelsom C, Wu X (2012) WOX2 and STIMPY-LIKE/WOX8 promote cotyledon boundary formation in Arabidopsis. Plant J 72:674–682. https://doi.org/10.1111/j.1365-313X.2012.05113.x
Liu YB, Lu SM, Zhang JF, Liu S, Lu YT (2007) A xyloglucan endotransglucosylase/hydrolase involves in growth of primary root and alters the deposition of cellulose in Arabidopsis. Planta 226:1547–1560. https://doi.org/10.1007/s00425-007-0591-2
Liu J, Sheng L, Xu Y, Li J, Yang Z, Huang H, Xu L (2014) WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26:1081–1093. https://doi.org/10.1105/tpc.114.122887
Liu K, Li Y, Chen X, Li L, Liu K, Zhao H, Wang Y, Han S (2018) ERF72 interacts with ARF6 and BZR1 to regulate hypocotyl elongation in Arabidopsis. J Exp Bot 69:3933–3947. https://doi.org/10.1093/jxb/ery220
Maris A, Suslov D, Fry SC, Verbelen JP, Vissenberg K (2009) Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J Exp Bot 60:3959–3972. https://doi.org/10.1093/jxb/erp229
Matsui A, Yokoyama R, Seki M, Ito T, Shinozaki K, Takahashi T, Komeda Y, Nishitani K (2005) AtXTH27 plays an essential role in cell wall modification during the development of tracheary elements. Plant J 42:525–534. https://doi.org/10.1111/j.1365-313X.2005.02395.x
Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95:805–815. https://doi.org/10.1016/S0092-8674(00)81703-1
Miedes E, Suslov D, Vandenbussche F, Kenobi K, Ivakov A, Van Der Straeten D, Lorences EP, Mellerowicz EJ, Verbelen JP, Vissenberg K (2013) Xyloglucan endotransglucosylase/hydrolase (XTH) overexpression affects growth and cell wall mechanics in etiolated Arabidopsis hypocotyls. J Exp Bot 64:2481–2497. https://doi.org/10.1093/jxb/ert107
Nakata M, Matsumoto N, Tsugeki R, Rikirsch E, Laux T, Okada K (2012) Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 24:519–535. https://doi.org/10.1105/tpc.111.092858
Ohmori Y, Tanaka W, Kojima M, Sakakibara H, Hirano HY (2013) WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in rice. Plant Cell 25:229–241. https://doi.org/10.1105/tpc.112.103432
Palovaara J, de Zeeuw T, Weijers D (2016) Tissue and organ initiation in the plant embryo: a first time for everything. Annu Rev Cell Dev Biol 32:47–75. https://doi.org/10.1146/annurev-cellbio-111315-124929
Park SO, Zheng Z, Oppenheimer DG, Hauser BA (2005) The PRETTY FEW SEEDS2 gene encodes an Arabidopsis homeodomain protein that regulates ovule development. Development 132:841–849. https://doi.org/10.1242/dev.01654
Penfield S (2017) Seed dormancy and germination. Curr Biol 27:R874–R878. https://doi.org/10.1016/j.cub.2017.05.050
Petzold HE, Chanda B, Zhao C, Rigoulot SB, Beers EP, Brunner AM (2018) DIVARICATA AND RADIALIS INTERACTING FACTOR (DRIF) also interacts with WOX and KNOX proteins associated with wood formation in Populus trichocarpa. Plant J 93:1076–1087. https://doi.org/10.1111/tpj.13831
Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, Groot E, Laux T (2015) Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev Cell 33:576–588. https://doi.org/10.1016/j.devcel.2015.04.024
Pillitteri LJ, Bemis SM, Shpak ED, Torii KU (2007) Haploinsufficiency after successive loss of signaling reveals a role for ERECTA-family genes in Arabidopsis ovule development. Development 134:3099–3109. https://doi.org/10.1242/dev.004788
Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, Job C, Job D (2012) Seed germination and vigor. Annu Rev Plant Biol 63:507–533. https://doi.org/10.1146/annurev-arplant-042811-105550
Romera-Branchat M, Ripoll JJ, Yanofsky MF, Pelaz S (2013) The WOX13 homeobox gene promotes replum formation in the Arabidopsis thaliana fruit. Plant J 73:37–49. https://doi.org/10.1111/tpj.12010
Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T (2007) Conserved factors regulate signaling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446:811–814. https://doi.org/10.1038/nature05703
Shu K, Liu XD, Xie Q, He ZH (2016) Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9:34–45. https://doi.org/10.1016/j.molp.2015.08.010
Singh AP, Tripathi SK, Nath P, Sane AP (2011) Petal abscission in rose is associated with the differential expression of two ethylene-responsive xyloglucan endotransglucosylase/hydrolase genes, RbXTH1 and RbXTH2. J Exp Bot 62:5091–5103. https://doi.org/10.1093/jxb/err209
Sliwinska E, Bassel GW, Bewley JD (2009) Germination of Arabidopsis thaliana seeds is not completed as a result of elongation of the radicle but of the adjacent transition zone and lower hypocotyl. J Exp Bot 60:3587–3594. https://doi.org/10.1093/jxb/erp203
Smit ME, McGregor SR, Sun H, Gough C, Bagman AM, Soyars CL, Kroon JT, Gaudinier A, Williams CJ, Yang XY, Nimchuk ZL, Weijers D, Turner SR, Brady SM, Etchells JP (2020) A PXY-mediated transcriptional network integrates signaling mechanisms to control vascular development in Arabidopsis. Plant Cell 32:319–335. https://doi.org/10.1105/tpc.19.00562
Steinbrecher T, Leubner-Metzger G (2017) The biomechanics of seed germination. J Exp Bot 68:765–783. https://doi.org/10.1093/jxb/erw428
Sun T, Goodman HM, Ausubel FM (1992) Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell 4:119–128. https://doi.org/10.1105/tpc.4.2.119
Sun B, Zhou YY, Cai J, Shang EL, Yamaguchi N, Xiao J, Looi LS, Wee WY, Gao XY, Wagner D, Ito T (2019) Integration of transcriptional repression and polycomb-mediated silencing of WUSCHEL in floral meristems. Plant Cell 31:1488–1505. https://doi.org/10.1105/tpc.18.00450
Tadege M, Lin H, Bedair M, Berbel A, Wen J, Rojas CM, Niu L, Tang Y, Sumner L, Ratet P, McHale NA, Madueno F, Mysore KS (2011) STENOFOLIA regulates blade outgrowth and leaf vascular patterning in Medicago truncatula and Nicotiana sylvestris. Plant Cell 23:2125–2142. https://doi.org/10.1105/tpc.111.085340
Tsuchiya M, Satoh S, Iwai H (2015) Distribution of XTH, expansin, and secondary-wall-related CesA in floral and fruit abscission zones during fruit development in tomato (Solanum lycopersicum). Front Plant Sci 6:323. https://doi.org/10.3389/fpls.2015.00323
Tvorogova VE, Krasnoperova EY, Potsenkovskaia EA, Kudriashov AA, Dodueva IE, Lutova LA (2021) What does the WOX say? Review of regulators, targets, partners. Mol Biol 55:311–337. https://doi.org/10.1134/S002689332102031X
Ueda M, Zhang Z, Laux T (2011) Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev Cell 20:264–270. https://doi.org/10.1016/j.devcel.2011.01.009
van der Graaff E, Laux T, Rensing SA (2009) The WUS homeobox-containing (WOX) protein family. Genome Biol 10:248. https://doi.org/10.1186/gb-2009-10-12-248
Van Sandt VS, Suslov D, Verbelen JP, Vissenberg K (2007) Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann Bot 100:1467–1473. https://doi.org/10.1093/aob/mcm248
Wang Z, Chen F, Li X, Cao H, Ding M, Zhang C, Zuo J, Xu C, Xu J, Deng X, Xiang Y, Soppe WJ, Liu Y (2016) Arabidopsis seed germination speed is controlled by SNL histone deacetylase-binding factor-mediated regulation of AUX1. Nat Commun 7:13412. https://doi.org/10.1038/ncomms13412
Wu X, Dabi T, Weigel D (2005) Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr Biol 15:436–440. https://doi.org/10.1016/j.cub.2004.12.079
Wu X, Chory J, Weigel D (2007) Combinations of WOX activities regulate tissue proliferation during Arabidopsis embryonic development. Dev Biol 309:306–316. https://doi.org/10.1016/j.ydbio.2007.07.019
Wu CC, Li FW, Kramer EM (2019) Large-scale phylogenomic analysis suggests three ancient superclades of the WUSCHEL-RELATED HOMEOBOX transcription factor family in plants. PLoS ONE 14:e0223521. https://doi.org/10.1371/journal.pone.0223521
Xie Q, Frugis G, Colgan D, Chua N-H (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14:3024–3036. https://doi.org/10.1101/gad.852200
Yasui Y, Ohmori Y, Takebayashi Y, Sakakibara H, Hirano HY (2018) WUSCHEL-RELATED HOMEOBOX4 acts as a key regulator in early leaf development in rice. PLoS Genet 14:e1007365. https://doi.org/10.1371/journal.pgen.1007365
Zhang Y, Jiao Y, Liu Z, Zhu YX (2015) ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells. Nat Commun 6:6003. https://doi.org/10.1038/ncomms7003
Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, Tarr PT, Yan A, Kay SA, Meyerowitz EM (2015) Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517:377–380. https://doi.org/10.1038/nature13853
Zhu J, Shi H, Lee BH, Damsz B, Cheng S, Stirm V, Zhu JK, Hasegawa PM, Bressan RA (2004) An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc Natl Acad Sci USA 101:9873–9878. https://doi.org/10.1073/pnas.0403166101
Zuo J, Niu QW, Frugis G, Chua NH (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30:349–359. https://doi.org/10.1046/j.1365-313X.2002.01289.x
Acknowledgements
The authors thank Dr. Galal Mabrouk very much for his great help improve the manuscript. This work was supported by grants from the National Natural Science Foundation of China (31870305, 32270353, and 32170355).
Author information
Authors and Affiliations
Contributions
YXL and HC planned and designed the research. YY, ZR, and LL conducted the experiments. YL and YH contributed materials and analysis tools. YY, ZR, YXL, and HC analyzed the data. YY and HC wrote the manuscript. YXL and HC revised the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest about the work described in this manuscript.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
425_2024_4357_MOESM1_ESM.docx
Supplementary file 1—Supplementary Fig. S1. Relative transcript abundance of the WOX family genes in dry, stratified, and germinating seeds, and seedlings. a–l The expression levels of WOX4, WOX6, WOX7, WOX5, WOX9, WOX11, WOX12, WOX13, WOX1, WOX2, WOX10, and WOX14 in dry, stratified, and germinating seeds, and seedlings. UBIQUITIN5 was used as an internal control for the normalization of gene expression. Values are means of three biological replicates. The error bars represent SE. Dry seeds, seeds stratified for 24 h, 48 h, and 72 h, seeds germinated for 8 h, 16 h, and seedlings germinated for 24 h, were collected for total RNA extraction. Fig. S2. Phylogenetic analysis of the Arabidopsis WOX family. The unrooted trees were inferred by the neighbor-joining method after the alignment of the protein sequences of the 15 Arabidopsis WOXs. WOX1, WOX2, and WOX4 are highlighted with the red box. Evolutionary analysis was conducted in the TBtools program. Fig. S3. Seed dormancy phenotypes of wild-type Col and wox mutants. a Germination after different dry storage periods of wild type (Col), wox2, and wox2-3. b Germination after different dry storage periods of Col, wox1, wox2, and wox1 wox2. c Germination after different dry storage periods of Col, wox2, wox4-1, and wox2 wox4-1. Seeds were stored at room temperature for after-ripening. Percentages of seed germination are means (± SE) based on at least eight individual plants for each genotype. Fig. S4. wox2 wox4-1 double mutant seedlings display a range of cotyledon phenotypes. a Six-day-old wild-type (Col) seedling with two normal cotyledons. b–e Six-day-old wox2 wox4-1 seedlings with two cotyledons positioned at an angle (b), two cotyledons of unequal size (c), single cotyledon (d) and heart-shaped cotyledon (e). Bars = 1 mm. Fig. S5. Total RNA was isolated from seeds imbibed for 18 h of wild type and wox2 wox4-1 for RNA-seq. Imbibition for 18 h was the first time point at which the germination of wox2 wox4-1 was significantly lower than that of Col. Seeds were stratified at 4 °C for 3 days before the germination experiment. Values are the means ± SE based on the seeds from six to eight individual plants for each genotype. The double asterisk indicates a significant difference compared to the wild type as determined by Student’s t-test (**P < 0.01). Fig. S6. Pathway enrichment analysis of DEGs in the boxes-marked pathways in Fig. 5a. XTHs genes are highlighted with the red box. The dot size is proportional to the gene number. The color scale indicates the significance level. (DOCX 1885 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, Y., Ren, Z., Li, L. et al. WOX2 functions redundantly with WOX1 and WOX4 to positively regulate seed germination in Arabidopsis. Planta 259, 83 (2024). https://doi.org/10.1007/s00425-024-04357-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-024-04357-7