Abstract
Main conclusion
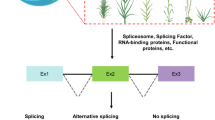
Serine/arginine-rich (SR) proteins participate in RNA processing by interacting with precursor mRNAs or other splicing factors to maintain plant growth and stress responses.
Abstract
Alternative splicing is an important mechanism involved in mRNA processing and regulation of gene expression at the posttranscriptional level, which is the main reason for the diversity of genes and proteins. The process of alternative splicing requires the participation of many specific splicing factors. The SR protein family is a splicing factor in eukaryotes. The vast majority of SR proteins’ existence is an essential survival factor. Through its RS domain and other unique domains, SR proteins can interact with specific sequences of precursor mRNA or other splicing factors and cooperate to complete the correct selection of splicing sites or promote the formation of spliceosomes. They play essential roles in the composition and alternative splicing of precursor mRNAs, providing pivotal functions to maintain growth and stress responses in animals and plants. Although SR proteins have been identified in plants for three decades, their evolutionary trajectory, molecular function, and regulatory network remain largely unknown compared to their animal counterparts. This article reviews the current understanding of this gene family in eukaryotes and proposes potential key research priorities for future functional studies.




Similar content being viewed by others
Data availability
Not applicable.
References
Albaqami M, Reddy ASN (2018) Development of an in vitro pre-mRNA splicing assay using plant nuclear extract. Plant Methods 14(1):1–12
Ali GS, Reddy AS (2006) ATP, phosphorylation and transcription regulate the mobility of plant splicing factors. J Cell Sci 119(Pt 17):3527–3538
Anko M (2014) Regulation of gene expression programmes by serine–arginine rich splicing factors. Semin Cell Dev Biol 32:11–21
Anko M, Mullermcnicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM (2012) The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol 13(3):1–17
Aravind L, Watanabe H, Lipman DJ, Koonin EV (2000) Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc Natl Acad Sci USA 97(21):11319–11324
Aznarez I, Nomakuchi TT, Tetenbaumnovatt J, Rahman MA, Fregoso O, Rees H, Krainer AR (2018) Mechanism of nonsense-mediated mRNA decay stimulation by splicing factor SRSF1. Cell Rep 23(7):2186–2198
Barta A, Kalyna M, Lorković ZJ (2008) Plant SR proteins and their functions. Curr Top Microbiol Immunol 326:83–102
Barta A, Kalyna M, Reddy AS (2010a) Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 22(9):2926–2929
Bedard KM, Daijogo S, Semler BL (2007) A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J 26(2):459–467
Blencowe BJ (2017) The relationship between alternative splicing and proteomic complexity. Trends Biochem Sci 42(6):407–408
Botti V, Mcnicoll F, Steiner MC, Richter FM, Solovyeva A, Wegener M, Schwich OD, Poser I, Zarnack K, Wittig I, Neugebauer KM, Müller-Mcnicoll M (2017) Cellular differentiation state modulates the mRNA export activity of SR proteins. J Cell Biol 216(7):1993–2009
Brugiolo M, Botti V, Liu N, Mullermcnicoll M, Neugebauer KM (2017) Fractionation iCLIP detects persistent SR protein binding to conserved, retained introns in chromatin, nucleoplasm and cytoplasm. Nucleic Acids Res 45(18):10452–10465
Busch A, Hertel KJ (2012) Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev Rna 3(1):1–12
Butt H, Bazin J, Prasad K, Awad N, Crespi M, Reddy ASN, Mahfouz MM (2022) The rice serine/arginine splicing factor RS33 regulates pre-mrna splicing during abiotic stress responses. Cells 11(11):1796
Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR (1997) Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol 138(2):225–238
Califice S, Baurain D, Hanikenne M, Motte P (2012a) A single ancient origin for prototypical serine/arginine-rich splicing factors. Plant Physiol 158(2):546–560
Chen T, Cui P, Xiong L (2015) The RNA-binding protein HOS5 and serine/arginine-rich proteins RS40 and RS41 participate in miRNA biogenesis in arabidopsis. Nucleic Acids Res 43(17):8283–8298
Chen Q, Han Y, Liu H, Wang X, Sun J, Zhao B, Li W, Tian J, Liang Y, Yan J, Yang X, Tian F (2018) Genome-wide association analyses reveal the importance of alternative splicing in diversifying gene function and regulating phenotypic variation in maize. Plant Cell 30(7):1404–1423
Chen MX, Wijethunge B, Zhou SM, Yang JF, Dai L, Wang SS, Chen C, Fu LJ, Zhang J, Hao GF, Yang GF (2019a) Chemical modulation of alternative splicing for molecular-target identification by potential genetic control in agrochemical research. J Agric Food Chem 67(18):5072–5084
Chen S, Li J, Liu Y, Li H (2019b) Genome-wide analysis of serine/arginine-rich protein family in wheat and brachypodium distachyon. Plants (basel) 8(7):188
Chen MX, Zhang KL, Zhang M, Das D, Fang YM, Dai L, Zhang J, Zhu FY (2020) Alternative splicing and its regulatory role in woody plants. Tree Physiol 40(11):1475–1486
Chen MX, Mei LC, Wang F, Boyagane Dewayalage IKW, Yang JF, Dai L, Yang GF, Gao B, Cheng CL, Liu YG, Zhang J, Hao GF (2021) PlantSPEAD: a web resource towards comparatively analysing stress-responsive expression of splicing-related proteins in plant. Plant Biotechnol J 19(2):227–229
Cho S, Hoang A, Sinha R, Zhong X, Fu X, Krainer AR, Ghosh G (2011) Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1–70K snRNP protein determines early spliceosome assembly. Proc Natl Acad Sci USA 108(20):8233–8238
Choi N, Liu Y, Oh J, Ha J, Ghigna C, Zheng X, Shen H (2021) Relative strength of 5’ splice-site strength defines functions of SRSF2 and SRSF6 in alternative splicing of Bcl-x pre-mRNA. BMB Rep 54(3):176–181
Dreyfuss G, Swanson MS, Piñol-Roma S (1988) Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem Sci 13(3):86–91
Duque P (2011) A role for SR proteins in plant stress responses. Plant Signal Behav 6(1):49–54
Fededa JP, Kornblihtt AR (2008) A splicing regulator promotes transcriptional elongation. Nat Struct Mol Biol 15(8):779–781
Feilner T, Hultschig C, Lee J, Meyer S, Immink RGH, Koenig A, Possling A, Seitz H, Beveridge A, Scheel D (2005) High throughput identification of potential arabidopsis mitogen-activated protein kinases substrates. Mol Cell Proteomics 4(10):1558–1568
Forment J, Naranjo MA, Roldán M, Serrano R, Vicente O (2002) Expression of arabidopsis SR-like splicing proteins confers salt tolerance to yeast and transgenic plants. Plant J 30(5):511–519
Fu X, Maniatis T (1990) Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature 343(6257):437–441
Giannakouros T, Nikolakaki E, Mylonis I, Georgatsou E (2011) Serine-arginine protein kinases: a small protein kinase family with a large cellular presence. FEBS J 278(4):570–586
Golovkin M, Reddy AS (1999) An SC35-like protein and a novel serine/arginine-rich protein interact with arabidopsis U1–70K protein. J Biol Chem 274(51):36428–36438
Graveley BR, Hertel KJ, Maniatis T (2001) The role of U2AF35 and U2AF65 in enhancer-dependent splicing. RNA 7(6):806–818
Grosse S, Lu YY, Coban I, Neumann B, Krebber H (2021) Nuclear SR-protein mediated mRNA quality control is continued in cytoplasmic nonsense-mediated decay. RNA Biol 18(10):1390–1407
Gu J, Ma S, Zhang Y, Wang D, Cao S, Wang ZY (2020) Genome-wide identification of cassava serine/arginine-rich proteins: insights into alternative splicing of pre-mrnas and response to abiotic stress. Plant Cell Physiol 61(1):178–191
Hartmann L, Wiesner T, Wachter A (2018a) Subcellular compartmentation of alternatively spliced transcripts defines SERINE/ARGININE-RICH PROTEIN30 expression. Plant Physiol 176(4):2886–2903
Hartmann L, Wießner T, Wachter A (2018b) Subcellular compartmentation of alternatively spliced transcripts defines SERINE/ARGININE-RICH PROTEIN30 expression. Plant Physiol 176(4):2886–2903
Hernandez H, Makarova OV, Makarov EM, Morgner N, Muto Y, Krummel DP, Robinson CV (2009) Isoforms of U1–70k control subunit dynamics in the human spliceosomal U1 snRNP. PLoS ONE 4(9):e7202
Holmes RK, Tuck AC, Zhu C, Dunn-Davies HR, Kudla G, Clauder-Munster S, Granneman S, Steinmetz LM, Guthrie C, Tollervey D (2015) Loss of the yeast SR protein Npl3 alters gene expression due to transcription readthrough. PLoS Genet 11(12):e1005735
Isshiki M, Tsumoto A, Shimamoto K (2006a) The serine/arginine-rich protein family in rice plays important roles in constitutive and alternative splicing of pre-mRNA. Plant Cell 18(1):146–158
Jeong S (2017) SR Proteins: binders, regulators, and connectors of RNA. Mol Cells 40(1):1–9
Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu X (2013) SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 153(4):855–868
Jimenez M, Urtasun R, Elizalde M, Azkona M, Latasa MU, Uriarte I, Arechederra M, Alignani D, Barcenavarela M, Alvarezsola G (2019) Splicing events in the control of genome integrity: role of SLU7 and truncated SRSF3 proteins. Nucleic Acids Res 47(7):3450–3466
Kalyna M, Lopato S, Barta A (2003) Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development. Mol Biol Cell 14(9):3565–3577
Kenan DJ, Query CC, Keene JD (1991) RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci 16(6):214–220
Kim K, Nguyen TD, Li S, Nguyen TA (2018) SRSF3 recruits DROSHA to the basal junction of primary microRNAs. RNA 24(7):892–898
Krainer AR, Maniatis T (1985) Multiple factors including the small nuclear ribonucleoproteins U1 and U2 are necessary for Pre-mRNA splicing in vitro. Cell 42(3):725–736
Laloum T, Martin G, Duque P (2017) Alternative splicing control of abiotic stress responses. Trends Plant Sci 23(2):140–150
Lareau LF, Inada M, Green RE, Wengrod J, Brenner SE (2007a) Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446(7138):926–929
Lazar G, Schaal TD, Maniatis T, Goodman HM (1995) Identification of a plant serine-arginine-rich protein similar to the mammalian splicing factor SF2/ASF. Proc Natl Acad Sci USA 92(17):7672–7676
Lemaire R, Prasad J, Kashima T, Gustafson J, Manley JL, Lafyatis R (2002) Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: a novel function for SR proteins. Genes Dev 16(5):594–607
Li X, Manley JL (2005) Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122(3):365–378
Lin S, Coutinhomansfield G, Wang D, Pandit S, Fu X (2008) The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol 15(8):819–826
Lopato S, Waigmann E, Barta A (1996) Characterization of a novel arginine/serine-rich splicing factor in arabidopsis. Plant Cell 8(12):2255–2264
Lorkovic ZJ, Hilscher J, Barta A (2008) Co-localisation studies of arabidopsis SR splicing factors reveal different types of speckles in plant cell nuclei. Exp Cell Res 314(17):3175–3186
Ma C, Ghosh G, Fu X, Adams JA (2010) Mechanism of dephosphorylation of the SR protein ASF/SF2 by protein phosphatase 1. J Mol Biol 403(3):386–404
Maertens GN, Cook N, Wang W, Hare S, Gupta SS, Oztop I, Lee K, Pye VE, Cosnefroy O, Snijders APL (2014) Structural basis for nuclear import of splicing factors by human transportin 3. Proc Natl Acad Sci USA 111(7):2728–2733
Manley JL, Krainer AR (2010) A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev 24(11):1073–1074
Maruyama K, Sato N, Ohta N (1999) Conservation of structure and cold-regulation of RNA-binding proteins in cyanobacteria: probable convergent evolution with eukaryotic glycine-rich RNA-binding proteins. Nucleic Acids Res 27(9):2029–2036
Meseguer S, Mudduluru G, Escamilla JM, Allgayer H, Barettino D (2011) MicroRNAs-10a and 10b contribute to retinoic acid-induced differentiation of neuroblastoma cells and target the alternative splicing regulatory factor SFRS1 (SF2/ASF). J Biol Chem 286(6):4150–4164
Michlewski G, Sanford JR, Caceres JF (2008) The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell 30(2):179–189
Morton M, Altamimi N, Butt H, Reddy ASN, Mahfouz M (2019a) Serine/Arginine-rich protein family of splicing regulators: new approaches to study splice isoform functions. Plant Sci 283:127–134
Mullermcnicoll M, Botti V, Domingues AMDJ, Brandl H, Schwich OD, Steiner MC, Curk T, Poser I, Zarnack K, Neugebauer KM (2016) SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev 30(5):553–566
Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, Obrien G, Shiue L, Clark TA, Blume JE, Ares M (2007) Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21(6):708–718
Palusa SG, Reddy ASN (2010) Extensive coupling of alternative splicing of pre-mRNAs of serine/arginine (SR) genes with nonsense-mediated decay. New Phytol 185(1):83–89
Palusa SG, Ali GS, Reddy AS (2007) Alternative splicing of pre-mRNAs of arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J 49(6):1091–1107
Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40(12):1413–1415
Reed R, Cheng H (2005) TREX, SR proteins and export of mRNA. Curr Opin Cell Biol 17(3):269–273
Richardson DN, Rogers MF, Labadorf A, Benhur A, Guo H, Paterson AH, Reddy ASN (2011) Comparative analysis of serine/arginine-rich proteins across 27 Eukaryotes: insights into sub-family classification and extent of alternative splicing. PLoS ONE 6(9):e24542
Rosenkranz RRE, Bachiri S, Vraggalas S, Keller M, Simm S, Schleiff E, Fragkostefanakis S (2021) Identification and regulation of tomato serine/arginine-rich proteins under high temperatures. Front Plant Sci 12:645689
Ruwe H, Kupsch C, Teubner M, Schmitz-Linneweber C (2011) The RNA-recognition motif in chloroplasts. J Plant Physiol 168(12):1361–1371
Sanford JR, Coutinho P, Hackett JA, Wang X, Ranahan WP, Caceres JF (2008a) Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS ONE 3(10):e3369
Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y (2008b) Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res 19(3):381–394
Savaldigoldstein S, Sessa G, Fluhr R (2000) The ethylene-inducible PK12 kinase mediates the phosphorylation of SR splicing factors. Plant J 21(1):91–96
Savaldigoldstein S, Aviv D, Davydov O, Fluhr R (2003) Alternative splicing modulation by a LAMMER kinase impinges on developmental and transcriptome expression. Plant Cell 15(4):926–938
Shi Y, Su Z, Yang H, Wang W, Jin G, He G, Siddique AN, Zhang L, Zhu A, Xue R, Zhang C (2019) Alternative splicing coupled to nonsense-mediated mRNA decay contributes to the high-altitude adaptation of maca (Lepidium meyenii). Gene 694:7–18
Stankovic N, Schloesser M, Joris M, Sauvage E, Hanikenne M, Motte P (2016) Dynamic distribution and interaction of the arabidopsis SRSF1 subfamily splicing factors. Plant Physiol 170(2):1000–1013
Sun S, Zhang Z, Sinha R, Karni R, Krainer AR (2010) SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol 17(3):306–312
Terzi LC, Simpson GG (2009) Arabidopsis RNA immunoprecipitation. Plant J 59(1):163–168
Tillemans V, Leponce I, Rausin G, Dispa L, Motte P (2006) Insights into nuclear organization in plants as revealed by the dynamic distribution of arabidopsis SR splicing factors. Plant Cell 18(11):3218–3234
Velazquezdones A, Hagopian JC, Ma C, Zhong X, Zhou H, Ghosh G, Fu X, Adams JA (2005) Mass spectrometric and kinetic analysis of ASF/SF2 phosphorylation by SRPK1 and Clk/Sty. J Biol Chem 280(50):41761–41768
Wan L, Deng M, Zhang H (2022) SR splicing factors promote cancer via multiple regulatory mechanisms. Genes (basel) 13(9):1659
Wang H, Jiang Y (2021) SRp20: A potential therapeutic target for human tumors. Pathol Res Pract 224:153444
Wang P, Xue L, Batelli G, Lee S, Hou Y, Van Oosten MJ, Zhang H, Tao WA, Zhu J (2013) Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc Natl Acad Sci USA 110(27):11205–11210
Wu JY, Maniatis T (1993) Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75(6):1061–1070
Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J (2010) A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell 38(1):67–77
Wu J, Liu H, Lu S, Hua J, Zou B (2021) Identification and expression analysis of chloroplast ribonucleoproteins (cpRNPs) in arabidopsis and rice. Genome 64(5):515–524
Xiao R, Sun Y, Ding J, Lin S, Rose DW, Rosenfeld MG, Fu X, Li X (2007) Splicing regulator sc35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol Cell Biol 27(15):5393–5402
Xiao R, Chen J, Liang Z, Luo D, Chen G, Lu ZJ, Chen Y, Zhou B, Li H, Du X (2019) Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell 178(1):107
Xing D, Wang Y, Hamilton M, Ben-Hur A, Reddy AS (2015) Transcriptome-wide identification of RNA targets of arabidopsis SERINE/ARGININE-RICH45 uncovers the unexpected roles of this RNA binding protein in RNA processing. Plant Cell 27(12):3294–3308
Xu S, Zhang Z, Jing B, Gannon P, Ding J, Xu F, Li X, Zhang Y (2011) Transportin-SR is required for proper splicing of resistance genes and plant immunity. PLoS Genet 7(6):e1002159
Yan Q, Xia X, Sun Z, Fang Y (2017) Depletion of Arabidopsis SC35 and SC35-like serine/arginine-rich proteins affects the transcription and splicing of a subset of genes. PLoS Genet 13(3):e1006663
Zhang Z, Krainer AR (2004) Involvement of SR proteins in mRNA Surveillance. Mol Cell 16(4):597–607
Zhang X, Shi Y, Powers JJ, Gowda NB, Zhang C, Ibrahim HM, Ball HB, Chen SL, Lu H, Mount SM (2017) Transcriptome analyses reveal SR45 to be a neutral splicing regulator and a suppressor of innate immunity in Arabidopsis thaliana. BMC Genomics 18(1):772–772
Zhao X, Tan L, Wang S, Shen Y, Guo L, Ye X, Liu S, Feng Y, Wu W (2021) The SR splicing factors: providing perspectives on their evolution, expression, alternative splicing, and function in populus trichocarpa. Int J Mol Sci 22(21):11369
Zhu FY, Chen MX, Ye NH, Shi L, Ma KL, Yang JF, Cao YY, Zhang Y, Yoshida T, Fernie AR, Fan GY, Wen B, Zhou R, Liu TY, Fan T, Gao B, Zhang D, Hao GF, Xiao S, Liu YG, Zhang J (2017a) Proteogenomic analysis reveals alternative splicing and translation as part of the abscisic acid response in arabidopsis seedlings. Plant J 91(3):518–533
Acknowledgements
This work was supported by the Guizhou Provincial Basic Research Program (Natural Science)-ZK[2023]-099, the National Natural Science Foundation of China (NSFC81401561, 91535109, 32001452), and the Hong Kong Research Grant Council (AoE/M-05/12, AoE/M-403/16, GRF14160516, 14177617, 12100318).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Institutional review board statement
Not applicable.
Informed consent statement
Not applicable.
Additional information
Communicated by Gerhard Leubner.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jia, ZC., Das, D., Zhang, Y. et al. Plant serine/arginine-rich proteins: versatile players in RNA processing. Planta 257, 109 (2023). https://doi.org/10.1007/s00425-023-04132-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-023-04132-0