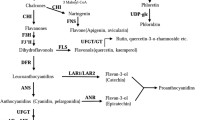
Abstract
Main conclusion
We identified 119 typical CaMYB encoding genes and reveal the major components of the proanthocyanidin regulatory network. CaPARs emerged as promising targets for genetic engineering toward improved agronomic traits in C. arietinum.
Abstract
Chickpea (Cicer arietinum) is among the eight oldest crops and has two main types, i.e., desi and kabuli, whose most obvious difference is the color of their seeds. We show that this color difference is due to differences in proanthocyanidin content of seed coats. Using a targeted approach, we performed in silico analysis, metabolite profiling, molecular, genetic, and biochemical studies to decipher the transcriptional regulatory network involved in proanthocyanidin biosynthesis in the seed coat of C. arietinum. Based on the annotated C. arietinum reference genome sequence, we identified 119 typical CaMYB encoding genes, grouped in 32 distinct clades. Two CaR2R3-MYB transcription factors, named CaPAR1 and CaPAR2, clustering with known proanthocyanidin regulators (PARs) were identified and further analyzed. The expression of CaPAR genes correlated well with the expression of the key structural proanthocyanidin biosynthesis genes CaANR and CaLAR and with proanthocyanidin levels. Protein–protein interaction studies suggest the in vivo interaction of CaPAR1 and CaPAR2 with the bHLH-type transcription factor CaTT8. Co-transfection analyses using Arabidopsis thaliana protoplasts showed that the CaPAR proteins form a MBW complex with CaTT8 and CaTTG1, able to activate the promoters of CaANR and CaLAR in planta. Finally, transgenic expression of CaPARs in the proanthocyanidin-deficient A. thaliana mutant tt2-1 leads to complementation of the transparent testa phenotype. Taken together, our results reveal main components of the proanthocyanidin regulatory network in C. arietinum and suggest that CaPARs are relevant targets of genetic engineering toward improved agronomic traits.








Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper and within the supplementary data published online.
Abbreviations
- ANR:
-
Anthocyanidin reductase
- BBS:
-
BHLH-binding sequences
- BiFC:
-
Bimolecular fluorescence complementation
- DMACA:
-
4-Dimethylaminocinnamaldehyde
- LAR:
-
Leucoanthocyanidin reductase
- MBS:
-
MYB-binding sequences
- PA:
-
Proanthocyanidin
- PAR:
-
Proanthocyanidin regulator
- TF:
-
Transcription factors
References
Akagi T, Katayama-Ikegami A, Kobayashi S et al (2012) Seasonal abscisic acid signal and a basic leucine zipper transcription factor, DkbZIP5, regulate proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol 158:1089–1102. https://doi.org/10.1104/pp.111.191205
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
An X-H, Tian Y, Chen K-Q et al (2015) MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol 56:650–662. https://doi.org/10.1093/pcp/pcu205
Appelhagen I, Thiedig K, Nordholt N et al (2014) Update on transparent testa mutants from Arabidopsis thaliana: characterisation of new alleles from an isogenic collection. Planta 240:955–970. https://doi.org/10.1007/s00425-014-2088-0
Bajaj D, Das S, Upadhyaya HD, Ranjan R, Badoni S, Kumar V, Tripathi S, Gowda CL, Sharma S, Singh S, Tyagi AK (2015) A genome-wide combinatorial strategy dissects complex genetic architecture of seed coat color in chickpea. Front Plant Sci 17:6–979. https://doi.org/10.3389/fpls.2015.00979
Bastian M, Heymann S, Jacomy M (2009) Gephi: an open source software for exploring and manipulating networks. Proc Intern AAAI Conference on Web and Social Media 3(1): 361–362. https://ojs.aaai.org/index.php/ICWSM/article/view/13937
Baudry A, Heim MA, Dubreucq B et al (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39:366–380. https://doi.org/10.1111/j.1365-313X.2004.02138.x
Bogs J, Jaffé FW, Takos AM et al (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143:1347–1361. https://doi.org/10.1104/pp.106.093203
Bondonno NP, Dalgaard F, Kyrø C et al (2019) Flavonoid intake is associated with lower mortality in the Danish diet cancer and health cohort. Nat Commun 10:3651. https://doi.org/10.1038/s41467-019-11622-x
Burbulis IE, Winkel-Shirley B (1999) Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc Natl Acad Sci USA 96:12929–12934. https://doi.org/10.1073/pnas.96.22.12929
Carmona-Gutierrez D, Zimmermann A, Kainz K et al (2019) The flavonoid 4,4’-dimethoxychalcone promotes autophagy-dependent longevity across species. Nat Commun 10:651. https://doi.org/10.1038/s41467-019-08555-w
Chang X, Xie S, Wei L et al (2020) Origins and stepwise expansion of R2R3-MYB transcription factors for the terrestrial adaptation of plants. Front Plant Sci 11:575360. https://doi.org/10.3389/fpls.2020.575360
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. https://doi.org/10.1046/j.1365-313x.1998.00343.x
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190. https://doi.org/10.1101/gr.849004
Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133:462–469. https://doi.org/10.1104/pp.103.027979
Deluc L, Barrieu F, Marchive C et al (2006) Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol 140:499–511. https://doi.org/10.1104/pp.105.067231
Deluc L, Bogs J, Walker AR et al (2008) The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol 147:2041–2053. https://doi.org/10.1104/pp.108.118919
Dobin A, Davis CA, Schlesinger F et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. https://doi.org/10.1093/bioinformatics/bts635
Dubos C, Stracke R, Grotewold E et al (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581. https://doi.org/10.1016/j.tplants.2010.06.005
Fan D, Wang X, Tang X et al (2018) Histone H3K9 demethylase JMJ25 epigenetically modulates anthocyanin biosynthesis in poplar. Plant J 96:1121–1136. https://doi.org/10.1111/tpj.14092
Garg R, Patel RK, Tyagi AK, Jain M (2011) De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res 18:53–63. https://doi.org/10.1093/dnares/dsq028
Gesell A, Yoshida K, Tran LT, Constabel CP (2014) Characterization of an apple TT2-type R2R3 MYB transcription factor functionally similar to the poplar proanthocyanidin regulator PtMYB134. Planta 240:497–511. https://doi.org/10.1007/s00425-014-2098-y
Haak M, Vinke S, Keller W et al (2018) High quality de novo transcriptome assembly of Croton tiglium. Front Mol Biosci 5:62. https://doi.org/10.3389/fmolb.2018.00062
Hartmann U, Sagasser M, Mehrtens F et al (2005) Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol 57:155–171. https://doi.org/10.1007/s11103-004-6910-0
He Q, Jones DC, Li W et al (2016) Genome-wide identification of R2R3-MYB genes and expression analyses during abiotic stress in Gossypium raimondii. Sci Rep 6:22980. https://doi.org/10.1038/srep22980
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300. https://doi.org/10.1093/nar/27.1.297
Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41:577–585. https://doi.org/10.1023/a:1006319732410
Katiyar A, Smita S, Lenka SK et al (2012) Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13:544. https://doi.org/10.1186/1471-2164-13-544
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Khandal H, Gupta SK, Dwivedi V et al (2020) Root-specific expression of chickpea cytokinin oxidase/dehydrogenase 6 leads to enhanced root growth, drought tolerance and yield without compromising nodulation. Plant Biotechnol J 18:2225–2240. https://doi.org/10.1111/pbi.13378
Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204:383–396. https://doi.org/10.1007/BF00331014
Koyama K, Numata M, Nakajima I et al (2014) Functional characterization of a new grapevine MYB transcription factor and regulation of proanthocyanidin biosynthesis in grapes. J Exp Bot 65:4433–4449. https://doi.org/10.1093/jxb/eru213
Kranz H, Scholz K, Weisshaar B (2000) c-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineages. Plant J 21:231–235. https://doi.org/10.1046/j.1365-313x.2000.00666.x
Kumar K, Purayannur S, Kaladhar VC et al (2018) mQTL-seq and classical mapping implicates the role of an AT-HOOK MOTIF containing nuclear localized (AHL) family gene in Ascochyta blight resistance of chickpea. Plant Cell Environ 41:2128–2140. https://doi.org/10.1111/pce.13177
Lavin M, Herendeen PS, Wojciechowski MF (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol 54:575–594. https://doi.org/10.1080/10635150590947131
Li C, Pei J, Yan X et al (2021) A poplar B-box protein PtrBBX23 modulates the accumulation of anthocyanins and proanthocyanidins in response to high light. Plant Cell Environ 44:3015–3033. https://doi.org/10.1111/pce.14127
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. https://doi.org/10.1093/bioinformatics/btt656
Liu X, Yu W, Zhang X et al (2017) Identification and expression analysis under abiotic stress of the R2R3-MYB genes in Ginkgo biloba L. Physiol Mol Biol Plants 23:503–516. https://doi.org/10.1007/s12298-017-0436-9
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Mao Z, Jiang H, Wang S et al (2021) The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple. Plant Sci 306:110848. https://doi.org/10.1016/j.plantsci.2021.110848
Marinova K, Pourcel L, Weder B et al (2007) The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+ -antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19:2023–2038. https://doi.org/10.1105/tpc.106.046029
Martin K, Kopperud K, Chakrabarty R et al (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59:150–162. https://doi.org/10.1111/j.1365-313X.2009.03850.x
Naik J, Rajput R, Pucker B et al (2021) The R2R3-MYB transcription factor MtMYB134 orchestrates flavonol biosynthesis in Medicago truncatula. Plant Mol Biol 106:157–172. https://doi.org/10.1007/s11103-021-01135-x
Nakagawa T, Kurose T, Hino T et al (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104:34–41. https://doi.org/10.1263/jbb.104.34
Nesi N, Jond C, Debeaujon I et al (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13:2099–2114. https://doi.org/10.1105/tpc.010098
Ogata K, Kanei-Ishii C, Sasaki M et al (1996) The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat Struct Biol 3:178–187. https://doi.org/10.1038/nsb0296-178
Pang Y, Peel GJ, Wright E et al (2007) Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol 145:601–615. https://doi.org/10.1104/pp.107.107326
Pang Y, Abeysinghe ISB, He J et al (2013) Functional characterization of proanthocyanidin pathway enzymes from tea and their application for metabolic engineering. Plant Physiol 161:1103–1116. https://doi.org/10.1104/pp.112.212050
Parween S, Nawaz K, Roy R et al (2015) An advanced draft genome assembly of a desi type chickpea (Cicer arietinum L.). Sci Rep 5:12806. https://doi.org/10.1038/srep12806
Pipas JM, Levine AJ (2001) Role of T antigen interactions with p53 in tumorigenesis. Semin Cancer Biol 11:23–30. https://doi.org/10.1006/scbi.2000.0343
Pucker B, Holtgräwe D, Rosleff Sörensen T et al (2016) A de novo genome sequence assembly of the Arabidopsis thaliana accession niederzenz-1 displays presence/absence variation and strong synteny. PLoS ONE 11:e0164321. https://doi.org/10.1371/journal.pone.0164321
Pucker B, Pandey A, Weisshaar B, Stracke R (2020) The R2R3-MYB gene family in banana (Musa acuminata): genome-wide identification, classification and expression patterns. PLoS ONE 15:e0239275. https://doi.org/10.1371/journal.pone.0239275
Quattrocchio F, Baudry A, Lepiniec L, Grotewold E (2006) The regulation of flavonoid biosynthesis. In: Grotewold E (ed) The science of flavonoids. Springer, New York, NY, pp 97–122
Rajkumar MS, Gupta K, Khemka NK, Garg R, Jain M (2020) DNA methylation reprogramming during seed development and its functional relevance in seed size/weight determination in chickpea. Commun Biol 3:1–3. https://doi.org/10.1038/s42003-020-1059-1
Rajput R, Naik J, Stracke R, Pandey A (2022) Interplay between R2R3 MYB-type activators and repressors regulates proanthocyanidin biosynthesis in banana (Musa acuminata). New Phytol. https://doi.org/10.1111/nph.18382
Ravaglia D, Espley RV, Henry-Kirk RA et al (2013) Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol 13:68. https://doi.org/10.1186/1471-2229-13-68
Routaboul J-M, Kerhoas L, Debeaujon I et al (2006) Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224:96–107. https://doi.org/10.1007/s00425-005-0197-5
Schwinn KE, Ngo H, Kenel F et al (2016) The onion (Allium cepa L.) R2R3-MYB gene MYB1 regulates anthocyanin biosynthesis. Front Plant Sci 7:1865. https://doi.org/10.3389/fpls.2016.01865
Seo J, Gordish-Dressman H, Hoffman EP (2006) An interactive power analysis tool for microarray hypothesis testing and generation. Bioinformatics 22:808–814
Shelton D, Stranne M, Mikkelsen L et al (2012) Transcription factors of lotus: regulation of isoflavonoid biosynthesis requires coordinated changes in transcription factor activity. Plant Physiol 159:531–547. https://doi.org/10.1104/pp.112.194753
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4:447–456
Stracke R, Ishihara H, Huep G et al (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50:660–677. https://doi.org/10.1111/j.1365-313X.2007.03078.x
Stracke R, Jahns O, Keck M et al (2010) Analysis of production of flavonol glycosides-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol 188:985–1000. https://doi.org/10.1111/j.1469-8137.2010.03421.x
Stracke R, Holtgräwe D, Schneider J et al (2014) Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biol 14:249. https://doi.org/10.1186/s12870-014-0249-8
Stracke R, Thiedig K, Kuhlmann M, Weisshaar B (2016) Analyzing synthetic promoters using Arabidopsis protoplasts. Methods Mol Biol 1482:67–81. https://doi.org/10.1007/978-1-4939-6396-6_5
Terrier N, Torregrosa L, Ageorges A et al (2009) Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol 149:1028–1041. https://doi.org/10.1104/pp.108.131862
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. https://doi.org/10.1093/nar/22.22.4673
Trezzini GF, Horrichs A, Somssich IE (1993) Isolation of putative defense-related genes from Arabidopsis thaliana and expression in fungal elicitor-treated cells. Plant Mol Biol 21:385–389. https://doi.org/10.1007/BF00019954
Vale M, Rodrigues J, Badim H et al (2021) Exogenous application of non-mature miRNA-encoded miPEP164c inhibits proanthocyanidin synthesis and stimulates anthocyanin accumulation in grape berry cells. Front Plant Sci 12:706679. https://doi.org/10.3389/fpls.2021.706679
Varma Penmetsa R, Carrasquilla-Garcia N, Bergmann EM et al (2016) Multiple post-domestication origins of kabuli chickpea through allelic variation in a diversification-associated transcription factor. New Phytol 211:1440–1451. https://doi.org/10.1111/nph.14010
Varshney RK, Song C, Saxena RK et al (2013) Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol 31:240–246. https://doi.org/10.1038/nbt.2491
Verdier J, Zhao J, Torres-Jerez I et al (2012) MtPAR MYB transcription factor acts as an on switch for proanthocyanidin biosynthesis in Medicago truncatula. Proc Natl Acad Sci USA 109:1766–1771. https://doi.org/10.1073/pnas.1120916109
Wang ZY, Kenigsbuch D, Sun L et al (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9:491–507. https://doi.org/10.1105/tpc.9.4.491
Wang N, Xu H, Jiang S et al (2017) MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J 90:276–292. https://doi.org/10.1111/tpj.13487
Wang N, Qu C, Jiang S et al (2018) The proanthocyanidin-specific transcription factor MdMYBPA1 initiates anthocyanin synthesis under low-temperature conditions in red-fleshed apples. Plant J 96:39–55. https://doi.org/10.1111/tpj.14013
Wilkins O, Nahal H, Foong J et al (2009) Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol 149:981–993. https://doi.org/10.1104/pp.108.132795
Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126:485–493. https://doi.org/10.1104/pp.126.2.485
Zhou H, Lin-Wang K, Liao L et al (2015) Peach MYB7 activates transcription of the proanthocyanidin pathway gene encoding leucoanthocyanidin reductase, but not anthocyanidin reductase. Front Plant Sci 6:908. https://doi.org/10.3389/fpls.2015.00908
Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40:22–34. https://doi.org/10.1111/j.1365-313X.2004.02183.x
Acknowledgements
This work was supported by the core grant of National Institute of Plant Genome Research and Department of Biotechnology grant (BT/PR36694/NNT/28/1722/2020) to AP. RR and JN acknowledge Council of Scientific and Industrial Research, Government of India, for Senior Research Fellowships. ST acknowledges Department of Biotechnology for Research Associate Fellowship. The authors are thankful to DBT-eLibrary Consortium (DeLCON) for providing access to e-resources. We acknowledge the Metabolome facility (BT/ INF/22/SP28268/2018) at NIPGR for phytochemical analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interest.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
425_2022_3979_MOESM1_ESM.tif
Supplementary file1 Chromosomal distribution of CaMYB genes. The localization of 108 R2R3-MYB genes are present on all eight pseudochromosomes in the C. arientinum genome; however, 11 CaMYB genes, i.e., CaMYB109 to CaMYB119, could not be assigned to any pseudochromosome. The pseudochromosomes of C. arientinum are shown with vertical bars and their chromosome numbers are presented at the top. The segmentally duplicated gene pair is highlighted in gray and connected with a black dotted line, while the tandemly duplicated gene pair is shown with green color. The length of each chromosome is given in Mb at its bottom (TIF 8999 KB)
425_2022_3979_MOESM2_ESM.tif
Supplementary file2 The conserved sequence logo of R2 and R3 MYB repeats of the 119 CaMYB proteins. Black triangles represent the conserved tryptophan (W) residues in the R2R3-MYB domain. Amino acid residues involved in the formation of conserved helices are indicated by black horizontal lines at the bottom of the logos (TIF 7535 KB)
425_2022_3979_MOESM3_ESM.pptx
Supplementary file3 Representative images of DMACA extracts of seed coats and cotyledons tissues for quantification of soluble and insoluble PAs in seed tissues of desi and kabuli types (PPTX 175 KB)
425_2022_3979_MOESM4_ESM.pptx
Supplementary file4 Ultra-high-performance liquid chromatography (UHPLC) chromatograms of standards and different tissues of desi and kabuli types. STD- Different peaks in the chromatogram are of procyanidin B1 (1); epigallocatechin (2); catechin (3); procyanidin B2 (4); procyanidin C1 (5); procyanidin A1 (6) epicatechin gallate (7); CA1, seed coat of ICC4958; CA2, seed coat of BGD256; CA3, seed coat of ICC6253; CA4, seed coat of ICCV2; CA5, cotyledon of ICC4958; CA6, cotyledon of BGD256; CA7, cotyledon of ICC6253; CA8, cotyledon of ICCV2 (PPTX 213 KB)
425_2022_3979_MOESM5_ESM.pptx
Supplementary file5 Liquid chromatography–mass spectrometry (LC-MS) chromatograms of epigallocatechin (EGC); epicatechin gallate (ECG); catechin (C); procyanidin A1 (PC-A1); procyanidin B1 (PC-B1); procyanidin B2 (PC-B2); procyanidin C1 (PC-C1) (PPTX 107 KB)
425_2022_3979_MOESM6_ESM.pptx
Supplementary file6 Representative images of DMACA extracts of PA-deficient A. thaliana tt2-1 mutant, the corresponding Ler wild type and transgenic 35S::CaPAR1, 2 lines in tt2-1 background (PPTX 274 KB)
425_2022_3979_MOESM7_ESM.tif
Supplementary file7 Phylogeny of TTG1 proteins. C. arietinum TTG1-like proteins (Ca_XP004502764.1, Ca_XP004502765, and Ca_XP004493117.1) were employed for the construction of phylogeny using neighbor Joining (NJ) method at 1000 bootstrap replication along with TTG1 proteins from Ananas comosus (OAY82628.1), Anthurium amnicola (JAT63430.1), Arabidopsis lyrata (XP_020877598.1), A. thaliana (CAB45372.1), Arabis alpina (KFK27728.1), Arachis duranensis (XP_015944381.1), Brassica napus (ABP04013.1), Brassica oleracea (ADV03945.1), Brassica rapa (ADK11704.1), Cajanus cajan (KYP38612.1), Camelina sativa (XP_010493614.1), Carica papaya (XP_021909514.1), Cucumis melo (XP_008455615.1), Daucus carota (XP_017254238.1), Dichanthelium oligosanthes (OEL12645.1), Eucalyptus grandis (XP_010048445.1), Gossypium arboreum (XP_017612811.1), G. hirsutum (AAM95642.1), Helianthus annuus (XP_021969841.1), Jatropha curcas (XP_012089919.1), Lotus corniculatus (ARK19314.1), L. sessilifolius (ATI97635.1), Malus domestica (ADI58760.1), Medicago truncatula (AES72643.1), Momordica charantia (XP_022140838.1), Morus notabilis (XP_010099415.1), Nelumbo lutea (ALU11273.1), Nicotiana attenuata (OIT04713.1), N. sylvestris (XP_009782290.1), N. tabacum (ACJ06978.1), Noccaea caerulescens (JAU90849.1), Oryza sativa (BAS80310.1), Prunus avium (XP_021805417), P. persica (ACQ65867.1), Punica granatum (ADV40946.1), Rosa rugosa (AFY23208.1), Setaria italica (XP_004953461.1), Sorghum bicolor (XP_002452749.2), Solanum melongena (AJN91103.1), Spinacia oleracea (XP_021858376.1), Triticum urartu (EMS66145.1), Zea mays (NP_001310302.1). Four major clades are shown in different colors, where Ca_XP004502764.1 and Ca_XP004502765.1 (blue highlighted) were closely clustered with TTG1 proteins from other leguminous plants (L. corniculatus, L. sessilifolius, C. cajan, A. duranensis, and M. truncatula), while Ca_XP004493117.1 (red highlighted) was found in a separate cluster with the members of TTG1 proteins from poaceae family including D. oligosanthes, O. sativa, S. italica, S. bicolor, and T. urartu (TIF 2251 KB)
425_2022_3979_MOESM8_ESM.tif
Supplementary file8 Co-transfection experiment in A. thaliana protoplasts. Results from co-transfection experiments in A. thaliana protoplasts. A GUS-fused 1328 bp CaANR and 1289 bp CaLAR promoter fragment (reporter) was assayed for its responsiveness to various 35S promoter-driven effectors CaMYB16, CaMYB30, CaTT8 and CaTTG1, either alone or in combinations. The figure shows mean normalized GUS' activity resulting from the influence of tested effector proteins on proCaANR-1328 and proCaLAR-1289 (reporter). Data from a set of six replicates are presented (TIF 2572 KB)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rajput, R., Tyagi, S., Naik, J. et al. The R2R3-MYB gene family in Cicer arietinum: genome-wide identification and expression analysis leads to functional characterization of proanthocyanidin biosynthesis regulators in the seed coat. Planta 256, 67 (2022). https://doi.org/10.1007/s00425-022-03979-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-022-03979-z