Abstract
Main conclusion
The oxidative pentose phosphate pathway provides cytosolic NADPH yet reduces carbon and energy use efficiency. Repressing this pathway and introducing cytosolic NADPH-dependent malate dehydrogenase may increase crop yields by ≈5%.
Abstract
Detailed knowledge about plant energy metabolism may aid crop improvements. Using published estimates of flux through central carbon metabolism, we phenotype energy metabolism in illuminated Camelina sativa leaves (grown at 22 °C, 500 µmol photons m−2 s−1) and report several findings. First, the oxidative pentose phosphate pathway (OPPP) transfers 3.3% of the NADPH consumed in the Calvin–Benson cycle to the cytosol. NADPH supply proceeds at about 10% of the rate of net carbon assimilation. However, concomitantly respired CO2 accounts for 4.8% of total rubisco activity. Hence, 4.8% of the flux through the Calvin–Benson cycle and photorespiration is spent on supplying cytosolic NADPH, a significant investment. Associated energy requirements exceed the energy output of the OPPP. Thus, autotrophic carbon metabolism is not simply optimised for flux into carbon sinks but sacrifices carbon and energy use efficiency to support cytosolic energy metabolism. To reduce these costs, we suggest bioengineering plants with a repressed cytosolic OPPP, and an inserted cytosolic NADPH-dependent malate dehydrogenase tuned to compensate for the loss in OPPP activity (if required). Second, sucrose cycling is a minor investment in overall leaf energy metabolism but a significant investment in cytosolic energy metabolism. Third, leaf energy balancing strictly requires oxidative phosphorylation, cofactor export from chloroplasts, and peroxisomal NADH import. Fourth, mitochondria are energetically self-sufficient. Fifth, carbon metabolism has an ATP/NADPH demand ratio of 1.52 which is met if ≤ 21.7% of whole electron flux is cyclic. Sixth, electron transport has a photon use efficiency of ≥ 62%. Last, we discuss interactions between the OPPP and the cytosolic oxidation–reduction cycle in supplying leaf cytosolic NADPH.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Central carbon metabolism operates co-ordinately across several cell compartments and requires compartment-specific inputs of reductant and energy cofactors (NAD(P)H, Fdred, ATP, UTP). These requirements vary with ambient and developmental conditions (Scheibe 2019). For instance, drought frequently causes stomatal closure and low intercellular CO2 concentrations which promote photorespiratory NADH production in mitochondria and consumption in peroxisomes (see below). Since cofactor production is spatially restricted, transmembrane transport and conversion of cofactors are key to metabolic functioning.
In illuminated leaves, reductant and energy cofactors are primarily produced by the light reactions of photosynthesis inside chloroplasts. Additionally, the pyruvate dehydrogenase complex (PDC) in fatty acid biosynthesis supplies chloroplastic NADH (Fig. 1). In the cytosol, glucose-6-phosphate dehydrogenase (G6PD) and 6-phosphogluconate dehydrogenase (6PGD) in the oxidative pentose phosphate pathway (OPPP), non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) in glycolysis, and isocitrate dehydrogenase (IDH) in amino acid biosynthesis supply NADPH while phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPC), phosphoglycerate kinase (PGK), and pyruvate kinase (PK) in glycolysis supply NADH and ATP. In mitochondria, the photorespiratory glycine decarboxylase complex (GDC) and several enzymes of the tricarboxylic acid cycle including PDC, and IDH supply NADH.
Sites of cofactor consumption and production in central carbon metabolism. Previously, we estimated carbon fluxes in 4-week-old photosynthesising Camelina sativa leaves (Xu et al. 2022). According to this analysis, pathways in black carry significant flux. Pathways in grey can be expected to carry significant flux but were not considered in our previous analysis. Enzymes: 6PGD, 6-phosphogluconate dehydrogenase; ACC, acetyl-CoA carboxylase; ACPr, 2,3-trans-enoyl-ACP reductase; AGPase, ADP-glucose pyrophosphorylase; FK, fructokinase; G6PD, glucose-6-phosphate dehydrogenase; GAPC (cytosolic) and GAPDH (chloroplastic), phosphorylating glyceraldehyde-3-phosphate dehydrogenase; GAPN, non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase; GDC, glycine decarboxylase complex; GDH, glutamate dehydrogenase; GK, glycerate kinase; GOGAT, glutamine-α-ketoglutarate aminotransferase; GS, glutamine synthetase; HK, hexokinase; HPR, hydroxypyruvate reductase; IDH, isocitrate dehydrogenase; KAR, 3-ketoacyl-ACP reductase; PDC, pyruvate dehydrogenase complex; PGK, phosphoglycerate kinase; PK, pyruvate kinase; PRK, phosphoribulokinase; UGPase, UDP-glucose pyrophosphorylase. Metabolites: 1,3BPG, 1,3-bisphosphoglycerate; 2PGA, 2-phosphoglycerate; 3PGA, 3-phosphoglycerate; 6PG, 6-phosphogluconate; 6PGL, 6-phosphogluconolactone; ACA, acetyl coenzyme A; ADPG, ADP-glucose; AKG, α-ketoglutarate; F6P, fructose 6-phosphate; FRU, fructose; G1P, glucose 1-phosphate; G6P, glucose 6-phosphate; GA, glycerate; GLC, glucose; GLN, glutamine; GLO, glyoxylate; GLU, glutamate; GLY, glycine; HP, hydroxypyruvate; ICT, isocitrate; PEP, phosphoenolpyruvate; PYR, pyruvate; Ru5P, ribulose 5-phosphate; RuBP, ribulose 1,5-bisphosphate; SER, serine; SUC, sucrose; TP, triose phosphate (glyceraldehyde 3-phosphate and dihydroxyacetone phosphate); UDPG, UDP-glucose
On the other hand, numerous enzyme reactions spread across several cell compartments consume reductant and energy cofactors. In chloroplasts, PGK, phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and phosphoribulokinase (PRK) in the Calvin–Benson cycle (CBC) consume NAD(P)H, and ATP. Furthermore, glutamine α-ketoglutarate aminotransferase (GOGAT), and glycerate kinase (GK) in photorespiration, ADP-glucose pyrophosphorylase (AGPase) in starch biosynthesis, and acetyl-CoA carboxylase (ACC), 3-ketoacyl-ACP reductase (KAR), and 2,3-trans-enoyl-ACP reductase (ACPr) in fatty acid biosynthesis consume chloroplastic NAD(P)H, Fdred, and ATP. In the cytosol, UDP-glucose pyrophosphorylase (UGPase) in sucrose biosynthesis, and hexokinase (HK), and fructokinase (FK) in sucrose cycling consume UTP, and ATP. In mitochondria, glutamine synthetase (GS) in photorespiration, and glutamate dehydrogenase (GDH) in amino acid biosynthesis consume ATP, and NADH while photorespiratory hydroxypyruvate reductase (HPR) consumes NADH in peroxisomes.
Several mechanisms enable transmembrane transport of cofactors from sites of production to sites of consumption (Fig. 2). NAD+ carrier proteins enable direct transport of NAD(P)H across the inner membranes of chloroplasts and mitochondria but have low affinities for these reductants (Palmieri et al. 2009). By contrast, dicarboxylate transporters coupled with malate dehydrogenase enzymes (malate valves) enable indirect exchange of NAD(P)H among chloroplasts, peroxisomes, mitochondria, and the cytosol (Selinski and Scheibe 2019). While there is an NADPH-dependent malate dehydrogenase inside chloroplasts, most malate dehydrogenases provide NADH, not NADPH. By contrast, glyceraldehyde 3-phosphate/3-phosphoglycerate (GAP/3PGA) cycling enables NADPH transfer from chloroplasts to the cytosol (Kelly and Gibbs 1973a, b). In this cycle, chloroplastic PGK and GAPDH reduce 3PGA to GAP consuming ATP and NADPH. Subsequently, GAP is exported to the cytosol where GAPN reduces it to 3PGA producing NADPH. A similar cycle involving GAPC, and PK (instead of GAPN) can provide cytosolic NADH and ATP at equimolar amounts (Stocking and Larson 1969). Additional cytosolic ATP can come from oxidative phosphorylation through the ADP/ATP carrier (Klingenberg 2008).
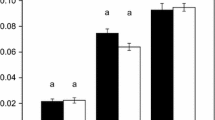
Compartment-specific cofactor consumption (negative values) and production (positive values) by central carbon metabolism in 4-week-old photosynthesising Camelina sativa leaves. The figure shows the net effect of all the processes listed in Table 2. Error bars represent 95% confidence intervals. 1, malate valves exchange NADH through malate-oxaloacetate interconversion by malate dehydrogenase (Selinski and Scheibe 2019). A chloroplastic isoform accepts NADPH. 2 and 3, cofactor transfer through redox cycles proposed by Kelly and Gibbs (1973a) and Stocking and Larson (1969), respectively. 4, counter-exchange of cytosolic ADP for mitochondrial ATP by the ADP/ATP carrier (Klingenberg 2008). 5, conversion of ATP and NADH to NADPH by the cytosolic oxidation–reduction cycle (Wieloch 2021). 6, NADH to ATP conversion by oxidative phosphorylation. 7, conversion of cytosolic NAD(P)H to mitochondrial ATP by oxidative phosphorylation starting at type II NAD(P)H dehydrogenases and glycerol-3-phosphate dehydrogenase both located in the inner mitochondrial membrane facing the cytosol. Solid arrow, direct cofactor transport. Dotted arrow, indirect cofactor transport via redox reactions
Furthermore, several mechanisms enable cofactor conversion. In the cytosol, equimolar amounts of ATP and NADH can be converted to NADPH by the carbon–neutral cytosolic oxidation–reduction (COR) cycle (Figs. 1, 2; Wieloch 2021). In its forward direction, cytosolic GAPN oxidises GAP to 3PGA producing NADPH. In its backward direction, cytosolic PGK and GAPC reduce 3PGA to GAP consuming ATP, and NADH. COR cycling may help balancing the cytosolic NADPH supply according to varying demands. For instance, we recently reported evidence for increased COR cycling under drought where increased synthesis of reactive oxygen species may require increased amounts of cytosolic NADPH to maintain the cellular redox balance (Wieloch et al. 2021). In mitochondria, NADH can be converted to ATP by oxidative phosphorylation. Similarly, cytosolic NAD(P)H can be converted to mitochondrial ATP by oxidative phosphorylation starting not at complex I but at type II NAD(P)H dehydrogenases or glycerol-3-phosphate dehydrogenase both located in the inner mitochondrial membrane facing the cytosol (Rasmusson et al. 2008).
In a recently published study, we estimated carbon fluxes across central metabolism in illuminated Camelina sativa leaves, including flux through the CBC, photorespiration, glycolysis, the tricarboxylic acid cycle, and starch, sucrose, fatty acid, and amino acid biosynthesis (Xu et al. 2022). Importantly, our analyses explained the 13C labelling lag of CBC metabolites (a longstanding conundrum in plant biochemistry) by reinjection of weakly labelled carbon from vacuolar and cytosolic sugar pools into the CBC. This process involves breakdown of sucrose into fructose and glucose, ATP-dependent re-phosphorylation to fructose 6-phosphate and glucose 6-phosphate, UTP-dependent sucrose resynthesis from fructose 6-phosphate and glucose 6-phosphate and reinjection of carbon from cytosolic glucose 6-phosphate into the CBC via the NADPH-producing OPPP (Fig. 1). Additionally, assimilation of the CO2 released by the OPPP involves all cofactor-dependent reactions in the CBC and photorespiration (ATP, NAD(P)H, Fdred).
Here, we comprehensively characterise energy metabolism in illuminated Camelina sativa leaves based on the carbon flux estimates of our previous study (Xu et al. 2022). More specifically, we first estimate compartment-specific cofactor demands of sucrose cycling and the OPPP and discuss associated effects on plant carbon and energy metabolism. Subsequently, we estimate compartment-specific cofactor demands of central carbon metabolism and address the following questions (with respect to the conditions and set of reactions studied here):
-
– Is cofactor export from chloroplasts strictly required?
-
– Are mitochondria energetically self–sufficient?
-
– Does photorespiration have the same chloroplastic cofactor requirements as the CBC?
-
– How much cyclic electron flux is required to meet the cofactor demands of carbon metabolism?
-
– Where is the cytosolic ATP coming from?
-
– Is oxidative phosphorylation strictly required? (Note that oxidative phosphorylation does not strictly require tricarboxylic acid cycling.)
-
– Is mitochondrial NADH strictly required to balance peroxisomal NADH demands?
-
– Where is the cytosolic NADPH coming from?
Materials and methods
Data
Previously, we estimated carbon fluxes across central metabolism in 4-week-old source leaves of wild-type Camelina sativa ecotype Suneson by 13C isotopically non-stationary metabolic flux analysis (Xu et al. 2022). These plants were grown at 22 °C, 50% relative humidity, and a light intensity of 500 µmol photons m−2 s−1 (8/16 h, day/night). Here, we utilise the carbon flux estimates to estimate associated cofactor fluxes (Fig. 1).
Cofactor demand by carbon metabolism
We resolved chloroplastic (.p), cytosolic (.c), mitochondrial (.m), and peroxisomal (.ox) fluxes. Within the Calvin–Benson cycle, we considered reactions catalysed by PGK.p (−1 ATP per turnover), GAPDH.p (−1 NAD(P)H per turnover), and PRK.p (−1 ATP per turnover). Within photorespiration, we considered reactions catalysed by GS.m (−1 ATP per turnover), GOGAT.p (−2 Fdred per turnover), GDC.m (+ 1 NADH per turnover), HPR.ox (−1 NADH per turnover), and GK.p (−1 ATP per turnover). It is commonly believed that photorespiratory ammonium is recaptured by GS2. While this enzyme is located in both chloroplasts and mitochondria (Taira et al. 2004), we here only considered the latter isoform since it has been suggested to immediately recapture potentially harmful ammonium released by GDC.m (Buchanan et al. 2015). Furthermore, while there is NADPH- and Fdred-dependent GOGAT.p, only the latter is essential for photorespiration and, therefore, considered here (Somerville and Ogren 1980). Within the OPPP, we considered reactions catalysed by G6PD.c (+ 1 NADPH per turnover), and 6PGD.c (+ 1 NADPH per turnover). Within starch and sucrose biosynthesis and sucrose cycling, we considered reactions catalysed by AGPase.p (−1 ATP per turnover), UGPase.c (−1 UTP per turnover), HK.c (−1 ATP per turnover), and FK.c (−1 ATP per turnover). Within glycolysis, we considered the reaction catalysed by PK.c (+ 1 ATP per turnover). Within the tricarboxylic acid cycle and amino acid biosynthesis, we only considered reactions catalysed by PDC.m (+ 1 NADH per turnover), IDH.m (+ 1 NADH per turnover), and GDH.m (−1 NADH per turnover) because other cofactor-dependent reactions reportedly carry negligible flux (Xu et al. 2022). Within fatty acid biosynthesis, we considered reactions catalysed by PDC.p (+ 1 NADH per turnover), ACC.p (−1 ATP per turnover), KAR.p (−1 NADPH per turnover), and ACPr.p (−1 NADH per turnover). Our accounting makes three assumptions. First, all pyruvate entering fatty acid biosynthesis comes from the β-elimination of phosphate from a carbocation intermediate of ribulose 1,5-bisphosphate (RuBP) carboxylation by rubisco (Andrews and Kane 1991). At 25 °C, the ratio of rubisco pyruvate synthesis to carboxylation is 0.7% (Andrews and Kane 1991). Xu et al. (2022) reported carboxylation proceeds at a rate of 172.1 µmol RuBP g−1 FW h−1 corresponding to ≈1.2 µmol g−1 FW h−1 pyruvate synthesis. Thus, pyruvate provided by rubisco significantly exceeds pyruvate requirements by fatty acid biosynthesis (0.4 µmol pyruvate g−1 FW h−1). Second, since the major isoform of ACPr.p is NADH-dependent and a minor isoform works with both NADH and NADPH (Buchanan et al. 2015), our accounting only considers NADH as cofactor of ACPr.p. Third, only C16 fatty acid are being produced. Initiation of C16 fatty acid biosynthesis requires 2 molecules pyruvate, 1 ATP, and 1 NADPH, and yields 1 C4 fatty acid, and 1 NADH. Elongation to a C16 fatty acid requires another 6 molecules pyruvate, 6 ATP, and 6 NADPH. Overall, biosynthesis of a C16 fatty acid requires 8 molecules pyruvate, 7 ATP, 7 NADPH and yields 1 NADH.
The cytosolic OPPP forms a shunt bypassing much of the CBC and releases CO2 from metabolism. We have calculated rubisco carboxylation and oxygenation rates accounting for the fixation of this CO2 as
and
where νOPPP denotes flux through the cytosolic OPPP, νc and νo denote rubisco carboxylation and oxygenation rates, respectively, Φ denotes the νo/νc ratio, and νc (OPPP) and νo (OPPP) denote rubisco carboxylation and oxygenation rates attributable to the CO2 lost in the OPPP, respectively. Please find the derivation of Eq. 1 in the supplement.
Results and discussion
Impacts of the cytosolic OPPP on plant carbon and energy metabolism
Flux through the cytosolic OPPP is likely a general feature of C3 metabolism since it can explain labelling lags of CBC metabolites (Xu et al. 2022) observed in several species fed 13CO2 or 14CO2 (Mahon et al. 1974; Canvin 1979; Hasunuma et al. 2010; Szecowka et al. 2013; Ma et al. 2014; Sharkey et al. 2020; Xu et al. 2021). In illuminated Camelina sativa leaves, the cytosolic OPPP supplies 14 µmol NADPH g−1 FW h−1, corresponding to 3.3% of the NAD(P)H consumed in the CBC (Tables 1, 2). NADPH supply proceeds at about 10% of the rate of net carbon assimilation. This NADPH may support cytosolic redox, signalling, and biosynthetic metabolism and mitochondrial ATP production (Rasmusson et al. 2008).
Cytosolic NADPH production by the OPPP is associated with the release of 7 μmol CO2 g−1 FW h−1 from metabolism (Table S1). Fixation of this CO2 accounts for 4.8% of the total activity of rubisco. That is, 4.8% of the total flux through the CBC and photorespiration is spent on providing cytosolic NADPH, a significant investment. Associated energy requirements (20 µmol NADPH g−1 FW h−1, 2.4 µmol Fdred g−1 FW h−1, 33 µmol ATP g−1 FW h−1) exceed the energy output of the OPPP (14 µmol NADPH g−1 FW h−1; Table 1). These carbon and energy costs would increase with increasing photorespiration. Thus, carbon metabolism in illuminated autotrophic tissue is not simply optimised for flux into carbon sinks but sacrifices both carbon and energy use efficiency to support cytosolic energy metabolism.
To try to reduce costs related to OPPP respiration, we suggest bioengineering and testing plants with a repressed cytosolic OPPP, and a newly introduced cytosolic NADPH-dependent malate dehydrogenase finely tuned to compensate for the loss in OPPP activity. This latter addition will only be required under conditions where carbon–neutral GAP/3PGA cycles and the COR cycle fall short of compensating for the reduction in NADPH from the OPPP.
Impact of sucrose cycling on plant energy metabolism
Sucrose cycling involving breakdown of sucrose into fructose and glucose, ATP-dependent re-phosphorylation to fructose 6-phosphate and glucose 6-phosphate, and UTP-dependent sucrose resynthesis from fructose 6-phosphate and glucose 6-phosphate is another futile carbon cycle (Fig. 1). In the cytosol of illuminated Camelina sativa leaves, its operation requires 6.5 µmol ATP g−1 FW h−1 (Table 1; phosphorylation of 1 UDP to UTP consumes 1 ATP). This corresponds to 1 and 53% of the ATP requirement in chloroplasts and the cytosol, respectively (Table 2). Thus, sucrose cycling is a minor investment in overall leaf energy metabolism but a significant investment in cytosolic energy metabolism. However, it is important to note that several processes consuming cytosolic ATP are not considered here including cytosol-vacuole proton pumping (Hedrich et al. 2015). Therefore, it is unlikely that half the ATP demand in the cytosol comes from sucrose cycling.
Cofactor fluxes in central carbon metabolism
Table 2 lists cofactor consumption and production by central enzyme reactions in carbon metabolism of Camelina sativa leaves. Figure 2 summarises this information for specific cell compartments. Chloroplast metabolism consumes large amounts of ATP and NADPH and some Fdred while producing small amounts of NADH. Cytosol metabolism consumes ATP and UTP while producing NADPH. If triose phosphates were the only cytosolic carbon substrates (i.e., if 3PGA was not exported) then there would be an additional supply of 2.15 (+ 0.19, − 0.16) µmol g−1 FW h−1 of NADH and ATP by GAPC and PGK or NADPH by GAPN (Fig. 1, grey reactions). Conversely, if exported 3PGA was reduced to triose phosphates, there would be an additional demand for cytosolic ATP and NADH by PGK and GAPC. However, these theoretical scenarios are not considered here since we have no estimates of carbon fluxes between cytosolic triose phosphates and 3PGA. Peroxisome metabolism consumes NADH, and mitochondrion metabolism consumes ATP while producing NADH. Compared to other cell compartments, absolute energy cofactor fluxes (ATP, UTP) in chloroplasts are 26.7-times higher while absolute reductant cofactor fluxes (NAD(P)H, Fdred) are 17.9-times higher.
Is cofactor export from chloroplasts strictly required?
Cofactor export from chloroplasts is mediated by the malate valve and GAP/3PGA cycles (Fig. 2). To investigate whether these processes are strictly required, we will now illustrate how cofactors produced by extra-chloroplastic carbon metabolism are used most efficiently to meet extra-chloroplastic cofactor demands. In this theoretical scenario, the mitochondrial NADH surplus (26.4 µmol g−1 FW h−1) is used to balance the peroxisomal NADH deficit (25.4 µmol g−1 FW h−1) through NADH transmembrane transport by malate valves (Fig. 2). Assuming a P/O ratio of 2.5 (Hinkle 2005), remaining mitochondrial NADH (1.03 µmol g−1 FW h−1) is converted to 2.6 µmol ATP g−1 FW h−1 via oxidative phosphorylation starting at complex I of the electron transport chain. Remaining mitochondrial and cytosolic ATP and UTP deficits equivalent to 34.1 µmol ATP g−1 FW h−1 are met by transmitting electrons from cytosolic NADPH onto the mitochondrial electron transport chain via type II NAD(P)H dehydrogenases. This reduces ATP yields by about 30% compared to transmission via complex I (Rasmusson et al. 2008). That is, the cytosolic surplus of 14 µmol NADPH g−1 FW h−1 is converted to 24.4 µmol ATP g−1 FW h−1 in mitochondria which is partly exported to the cytosol via the ADP/ATP carrier (Klingenberg 2008). Overall, there remains an extra-chloroplastic deficit of 9.7 µmol ATP g−1 FW h−1. Thus, with respect to the conditions and set of reactions studied here, cofactor export from chloroplasts is strictly required to meet all extra-chloroplastic cofactor demands.
Are mitochondria energetically self-sufficient?
It may be most realistic to assume that the mitochondrial surplus of 26.4 µmol NADH g−1 FW h−1 is used first to balance the mitochondrial deficit of 25.5 µmol ATP g−1 FW h−1 rather than being directly shuttled out of mitochondria into peroxisomes (Fig. 2). To meet the mitochondrial ATP deficit by oxidative phosphorylation starting from complex I requires only 10.2 µmol NADH g−1 FW h−1. Thus, with respect to the conditions and set of reactions studied here, mitochondria are energetically self-sufficient.
Does photorespiration have the same chloroplastic cofactor requirements as the CBC?
Our cofactor accounting assumes that photorespiratory ammonium is recaptured by mitochondrial (not chloroplastic) GS. This has implications for the cofactor requirements in chloroplasts. Chloroplastic cofactor requirements upon carboxylation of 2 RuBP comprise 4 ATP by PGK, 4 NADPH by GAPDH, and 2 ATP by PRK. Chloroplastic cofactor requirements upon oxygenation of 2 RuBP comprise 3 ATP by PGK, 3 NADPH by GAPDH, 2 ATP by PRK, 1 ATP by GK, and 2 Fdred corresponding to 1 NADPH by GOGAT. Thus, overall, both processes have the same absolute chloroplastic cofactor requirements with a chloroplastic ATP/NADPH demand ratio of 1.5. Other processes, such as starch biosynthesis and serine withdrawal from photorespiration, will cause only small changes of this ratio (see below).
How much cyclic electron flux is required to meet the cofactor demands of carbon metabolism?
Chloroplast carbon metabolism has much higher demands for reductant and energy cofactors than any other cell compartment (Fig. 2). These demands are readily satisfied by cofactor input mainly from the light reactions of photosynthesis without the need for transmembrane transport or conversion of cofactors. Interestingly, chloroplast GAPDH accepts both NADPH and NADH (McGowan and Gibbs 1974). In vivo, however, NADPH supply from the light reactions is 7630-times higher than the NADH surplus from fatty acid biosynthesis (Table 2).
Overall, chloroplast metabolism requires 680 µmol ATP g−1 FW h−1 and 446 µmol NAD(P)H g−1 FW h−1 (Table 2). This accounts for Fdred demands where 2 Fdred correspond to 1 NADPH. Hence, the reaction network and conditions studied here have an ATP/NADPH demand ratio of 1.52. However, linear electron flux including Q cycling has an estimated ATP/NADPH supply ratio of 1.286 (assuming trans-membrane transport of 3 protons per electron and production of 3 ATP per 14 protons by ATP synthase) (Seelert et al. 2000). Thus, assuming neither the water-water cycle (affecting the ATP/NADPH supply ratio) nor GAP/3PGA cycles and the chloroplast malate valve (affecting the ATP/NADPH demand ratio) are operational, cyclic electron flux through photosystem I is required.
Production of 446 µmol NADPH g−1 FW h−1 requires linear electron flux at a rate of 892 µmol g−1 FW h−1 and yields 574 µmol ATP g−1 FW h−1. To meet the remaining deficit of 106 µmol ATP g−1 FW h−1, cyclic electron flux through photosystem I including Q cycling at a rate of 248 µmol g−1 FW h−1 is required (assuming trans-membrane transport of 2 protons per electron and production of 3 ATP per 14 protons by ATP synthase) (Seelert et al. 2000). Thus, with respect to the conditions and set of reactions studied here, cyclic electron flux accounts for 21.7% of whole electron flux and is a 27.8% fraction of linear electron flux.
Jointly, linear, and cyclic electron flux through photosystem I consume 2032 µmol photons g−1 FW h−1. Carbon flux in Camelina sativa leaves was measured at 500 µmol photons m−2 s−1 corresponding to 3273 µmol photons g−1 FW h−1 (the fresh weight per leaf area for a typical Camelina leaf is 0.055 ± 0.0041 g cm−2 implying a leaf thickness of 0.55 mm). Thus, with respect to the conditions and set of reactions studied here, 62% of the photons falling on the leaf are absorbed and used for photosynthesis.
In our previous analysis (Xu et al. 2022), carbon fluxes through GAP/3PGA cycles, the COR cycle (grey in Fig. 1), and the chloroplast malate valve were not estimated, i.e., associated cofactor fluxes are unknown. These processes would, however, decrease the ATP/NADPH demand ratio of carbon metabolism and, thus, the requirement for cyclic electron flux. This is because GAP/3PGA cycles require chloroplastic ATP and NAD(P)H at a 1:1 ratio (Stocking and Larson 1969; Kelly and Gibbs 1973a), and the malate valve requires chloroplastic NAD(P)H only (Selinski and Scheibe 2019). COR cycling requires NADH and ATP in the cytosol which may be provided by combined action of the chloroplast malate valve and oxidative phosphorylation (Wieloch 2021). Furthermore, these processes would consume additional electrons and, thus, increase the photon use efficiency of electron transport. For example, no cyclic electron flux is required if 82.5 µmol NAD(P)H g−1 FW h−1 (corresponds to ≈60% of net CO2 assimilation) are exported by the chloroplast malate valve. This additional flux would result in 64.6% photon use efficiency of electron transport.
Where is the cytosolic ATP coming from? Is oxidative phosphorylation strictly required?
Overall, we estimated an extra-chloroplastic ATP (includes UTP) deficit of 36.7 µmol g−1 FW h−1 (Fig. 2, Table 2) which may be balanced by the following mechanisms. First, cytosolic NADPH from the OPPP (14 µmol g−1 FW h−1) may be converted to 24.4 µmol ATP g−1 FW h−1 via oxidative phosphorylation starting at type II NADPH dehydrogenase (yields 2.5 × 0.7 ATP per NADPH). This leaves an ATP deficit of 12.3 µmol g−1 FW h−1. Second, GAP/3PGA cycling involving cytosolic GAPN may supply cytosolic NADPH (Fig. 1). To balance the entire ATP deficit of 36.7 µmol g−1 FW h−1 via oxidative phosphorylation starting at type II NADPH dehydrogenases would require a flux of 21 µmol GAP g−1 FW h−1 through this pathway. Third, mitochondrial NADH from photorespiration (26.4 µmol g−1 FW h−1) may be converted to up to 66 µmol ATP g−1 FW h−1 via oxidative phosphorylation starting at complex I of the mitochondrial electron transport chain (yields 2.5 ATP per NADH). Fourth, GAP/3PGA cycling involving cytosolic PGK may supply ATP (Fig. 1). This pathway provides ATP and NADH (from GAPC) at equimolar amounts. If the latter is imported into mitochondria by the malate valve and used for oxidative phosphorylation (Fig. 2), GAP/3PGA cycling at a rate of 10.5 µmol g−1 FW h−1 would suffice to meet the ATP deficit of 36.7 µmol g−1 FW h−1 (1 ATP from PGK, 2.5 ATP from NADH provided by GAPC). If NADH from GAPC enters oxidative phosphorylation via type II NADH dehydrogenase which reduces ATP yields by about 30% (see above), GAP/3PGA cycling at a rate of 13.3 µmol g−1 FW h−1 is required to meet the ATP deficit (1 ATP from PGK, 2.5 × 0.7 ATP from NADH). Fifth, GAP/3PGA cycling involving cytosolic PGK may supply all extra-chloroplastic ATP required (Fig. 1). This would provide an additional 36.7 µmol NADH g−1 FW h−1 from GAPC which could be used to balance the peroxisomal NADH deficit (25.4 µmol g−1 FW h−1). Remaining extra-chloroplastic NADH (37.7 µmol g−1 FW h−1, includes the mitochondrial NADH surplus) would then most likely enter oxidative phosphorylation. Sixth, cytosolic NADH supplied by the chloroplast malate valve may be converted to ATP by mitochondrial NADH import (malate valve) and oxidative phosphorylation or oxidative phosphorylation via type II NADH dehydrogenase. Supply of 36.7 µmol ATP g−1 FW h−1 by these pathways requires net flux through the chloroplast malate valve of 14.7 or 21 µmol NADH g−1 FW h−1, respectively. Hence, numerous metabolic pathways can supply cytosolic ATP. All of them, however, involve oxidative phosphorylation. Thus, in illuminated Camelina sativa leaves under the conditions studied here, oxidative phosphorylation is strictly required. This is in agreement with results from a recent simulation study (Shameer et al. 2019).
Is mitochondrial NADH strictly required to balance peroxisomal NADH demands?
We estimated a peroxisomal NADH deficit of 25.4 µmol g−1 FW h−1 due to photorespiration (Fig. 2, Table 2). This deficit is often assumed to be balanced by the mitochondrial NADH surplus of 26.4 µmol g−1 FW h−1 coming mostly from photorespiration. However, there are other processes that may help to balance the peroxisomal NADH deficit. First, NAD(P)H from chloroplasts may be transferred to peroxisomes via malate valves. Second, combined action of GAP/3PGA cycling involving cytosolic GAPC and the peroxisomal malate valve may balance peroxisomal NADH demands. Concomitantly supplied ATP by PGK may help to balance extra-chloroplastic ATP deficits. Thus, to balance peroxisomal NADH demands, mitochondrial NADH is not strictly required. This is in agreement with results from a recent simulation study (Shameer et al. 2019). However, peroxisomal NADH import appears to be strictly required.
Where is the cytosolic NADPH coming from?
In the cytosol of illuminated Camelina sativa leaves, 14 µmol NADPH g−1 FW h−1 are supplied by the OPPP. Primary flux control through the OPPP is exerted at the level of its first enzyme, G6PD. G6PD activity in Solanum tuberosum leaf discs was found to be constant in response to increasing H2O2 (Hauschild and von Schaewen 2003). By contrast, increases in activity occur with increasing sugar availability (sugars tested: mannose, glucose, fructose, sucrose) through de novo enzyme synthesis (Hauschild and von Schaewen 2003). As carbon assimilation decreases, so does sugar availability (Lawlor and Fock 1977; Sánchez-Rodríguez et al. 1999). This can be expected to result in decreasing OPPP flux and associated NADPH supply. However, decreasing carbon assimilation (e.g., due to drought) also promotes the synthesis of reactive oxygen species (Noctor et al. 2002) and their detoxification can be expected to increase the demand for cytosolic NADPH. Thus, especially (but not exclusively) under low carbon assimilation, other sources of cytosolic NADPH may be required.
GAPN catalyses the irreversible oxidation of GAP to 3PGA and simultaneous reduction of NADP+ to NADPH (Fig. 1). Interestingly, a GAPN null mutant showed increased expression and activity of cytosolic G6PD (3.8-fold increase in At3g27300, 3.2-fold increase in At5g40760) and an increased oxidative load (Rius et al. 2006). This suggests that G6PD and GAPN interact in redox control and that GAPN is required in redox control since increased G6PD expression cannot fully compensate for the loss in GAPN function.
Just like GAPN, GAPC and PGK catalyse cytosolic GAP to 3PGA conversions. Intriguingly, the GAPN null mutant showed increased expression and activity of GAPC (GAPC1: 3.5-fold increase in At3g04120) (Rius et al. 2006). This was suggested to compensate for the lack of GAPN activity (Rius et al. 2006). However, GAPC and PGK supply NADH and ATP which (in contrast to NADPH) cannot be used in redox control. Alternatively, upregulation of GAPC may promote COR cycling (Wieloch 2021). Each turn of this cycle provides NADPH at GAPN and consumes ATP and NADH at PGK and GAPC (Fig. 1). Required NADH and ATP may be provided via the chloroplast malate valve and oxidative phosphorylation (Fig. 2). In this regard, a putative moonlighting function of GAPC is worth noting (Scheibe 2019). With increasing oxidative load (as observed in the GAPN null mutant), GAPC is increasingly deactivated and moved to the nucleus (Piattoni et al. 2013; Hildebrandt et al. 2015). There, it can bind to a gene coding for chloroplast malate dehydrogenase, and expression of this gene was found to increase during the initial stages of oxidative stress (Hildebrandt et al. 2015). Furthermore, it is worth noting that the GAPN null mutant shows increased expression of cytosolic malate dehydrogenase (Rius et al. 2006). Taken together, this may result in increased malate valve capacity to support GAPC and PGK in COR cycling.
The most marked difference between cytosolic NADPH supply by the OPPP versus COR cycling is that the latter does not release CO2. Thus, cytosolic NADPH supply by COR cycling might be physiologically beneficial when CO2 assimilation is impeded, e.g., under photorespiratory conditions. In line with this reasoning, we recently reported isotope evidence for upregulation of COR cycling under drought (Wieloch et al. 2021). Under these same conditions, regulatory properties of G6PD suggest decreased flux through the OPPP (see above). Taken together, we propose that the OPPP and COR cycling act in concert to ensure the cytosolic NADPH supply under varying environmental/developmental conditions. To follow up on this proposal, wide-screening enzyme expression in a G6PD knockout mutant may be valuable.
Author contribution statement
TW and TDS designed the research. TW led the research. TW analysed and interpreted the data and wrote the manuscript with input from TDS.
Data availability
All data generated or analysed during this study are included in this published article.
References
Andrews TJ, Kane HJ (1991) Pyruvate is a by-product of catalysis by ribulosebisphosphate carboxylase/oxygenase. J Biol Chem 266:9447–9452
Buchanan BB, Gruissem W, Jones RL (eds) (2015) Biochemistry and molecular biology of plants, 2nd edn. John Wiley and Sons, Ltd, Chichester, UK
Canvin DT (1979) Photorespiration: comparison between C3 and C4 plants. In: Gibbs M, Latzko E (eds) Photosynthesis II: photosynthetic carbon metabolism and related processes. Springer, Berlin Heidelberg, Berlin, Germany, pp 368–396
Hasunuma T, Harada K, Miyazawa S-I et al (2010) Metabolic turnover analysis by a combination of in vivo 13C-labelling from 13CO2 and metabolic profiling with CE-MS/MS reveals rate-limiting steps of the C3 photosynthetic pathway in Nicotiana tabacum leaves. J Exp Bot 61:1041–1051
Hauschild R, von Schaewen A (2003) Differential regulation of glucose-6-phosphate dehydrogenase isoenzyme activities in potato. Plant Physiol 133:47–62
Hedrich R, Sauer N, Neuhaus HE (2015) Sugar transport across the plant vacuolar membrane: nature and regulation of carrier proteins. Curr Opin Plant Biol 25:63–70
Hildebrandt T, Knuesting J, Berndt C et al (2015) Cytosolic thiol switches regulating basic cellular functions: GAPDH as an information hub? Biol Chem 396:523–537
Hinkle PC (2005) P/O ratios of mitochondrial oxidative phosphorylation. Biochim Biophys Acta Bioenerg 1706:1–11
Kelly GJ, Gibbs M (1973a) Nonreversible D-glyceraldehyde 3-phosphate dehydrogenase of plant tissues. Plant Physiol 52:111–118
Kelly GJ, Gibbs M (1973b) A mechanism for the indirect transfer of photosynthetically reduced nicotinamide adenine dinucleotide phosphate from chloroplasts to the cytoplasm. Plant Physiol 52:674–676
Klingenberg M (2008) The ADP and ATP transport in mitochondria and its carrier. Biochim Biophys Acta Biomembr 1778:1978–2021
Lawlor DW, Fock H (1977) Water stress induced changes in the amounts of some photosynthetic assimilation products and respiratory metabolites of sunflower leaves. J Exp Bot 28:329–337
Ma F, Jazmin LJ, Young JD, Allen DK (2014) Isotopically nonstationary 13C flux analysis of changes in Arabidopsis thaliana leaf metabolism due to high light acclimation. Proc Natl Acad Sci USA 111:16967–16972
Mahon JD, Fock H, Canvin DT (1974) Changes in specific radioactivity of sunflower leaf metabolites during photosynthesis in 14CO2 and 12CO2 at three concentrations of CO2. Planta 120:245–254
McGowan RE, Gibbs M (1974) Comparative enzymology of the glyceraldehyde 3-phosphate dehydrogenases from Pisum sativum. Plant Physiol 54:312
Noctor G, Veljovic-Jovanovic S, Driscoll S et al (2002) Drought and oxidative load in the leaves of C3 plants: a predominant role for photorespiration? Ann Bot 89:841–850
Palmieri F, Rieder B, Ventrella A et al (2009) Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins. J Biol Chem 284:31249–31259
Piattoni CV, Guerrero SA, Iglesias AA (2013) A differential redox regulation of the pathways metabolizing glyceraldehyde-3-phosphate tunes the production of reducing power in the cytosol of plant cells. Int J Mol Sci 14:8073–8092
Rasmusson AG, Geisler DA, Moller IM (2008) The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion 8:47–60
Rius SP, Casati P, Iglesias AA, Gomez-Casati DF (2006) Characterization of an Arabidopsis thaliana mutant lacking a cytosolic non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase. Plant Mol Biol 61:945–957
Sánchez-Rodríguez J, Pérez P, Martínez-Carrasco R (1999) Photosynthesis, carbohydrate levels and chlorophyll fluorescence-estimated intercellular CO2 in water-stressed Casuarina equisetifolia Forst. and Forst. Plant Cell Env 22:867–873
Scheibe R (2019) Maintaining homeostasis by controlled alternatives for energy distribution in plant cells under changing conditions of supply and demand. Photosynth Res 139:81–91
Seelert H, Poetsch A, Dencher NA et al (2000) Proton-powered turbine of a plant motor. Nature 405:418–419
Selinski J, Scheibe R (2019) Malate valves: old shuttles with new perspectives. Plant Biol 21:21–30
Shameer S, Ratcliffe RG, Sweetlove LJ (2019) Leaf energy balance requires mitochondrial respiration and export of chloroplast NADPH in the light. Plant Physiol 180:1947–1961
Sharkey TD, Preiser AL, Weraduwage SM, Gog L (2020) Source of 12C in calvin-benson cycle intermediates and isoprene emitted from plant leaves fed with 13CO2. Biochem J 477:3237–3252
Somerville CR, Ogren WL (1980) Inhibition of photosynthesis in Arabidopsis mutants lacking leaf glutamate synthase activity. Nature 286:257–259
Stocking CR, Larson S (1969) A chloroplast cytoplasmic shuttle and the reduction of extraplastid NAD. Biochem Biophys Res Commun 37:278–282
Szecowka M, Heise R, Tohge T et al (2013) Metabolic fluxes in an illuminated Arabidopsis rosette. Plant Cell 25:694–714
Taira M, Valtersson U, Burkhardt B, Ludwig RA (2004) Arabidopsis thaliana GLN2-encoded glutamine synthetase is dual targeted to leaf mitochondria and chloroplasts. Plant Cell 16:2048–2058
Wieloch T (2021) A cytosolic oxidation–reduction cycle in plant leaves. J Exp Bot 72:4186–4189
Wieloch T, Werner RA, Schleucher J (2021) Carbon flux around leaf-cytosolic glyceraldehyde-3-phosphate dehydrogenase introduces a 13C signal in plant glucose. J Exp Bot 72:7136–7144
Xu Y, Fu X, Sharkey TD et al (2021) The metabolic origins of non-photorespiratory CO2 release during photosynthesis: a metabolic flux analysis. Plant Physiol 186:297–314
Xu Y, Wieloch T, Kaste JAM et al (2022) Reimport of carbon from cytosolic and vacuolar sugar pools into the calvin-benson cycle explains photosynthesis labeling anomalies. PNAS 119:e2121531119
Acknowledgements
T.D.S. was supported by the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the United States Department of Energy Grant DE-FG02-91ER20021. T.D.S. receives partial salary support from MSU AgBioResearch. However, the contents of this paper are solely the responsibility of the authors.
Funding
Open access funding provided by Umea University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wieloch, T., Sharkey, T.D. Compartment-specific energy requirements of photosynthetic carbon metabolism in Camelina sativa leaves. Planta 255, 103 (2022). https://doi.org/10.1007/s00425-022-03884-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-022-03884-5