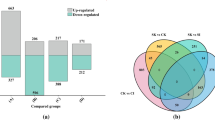
Abstract
Main conclusion
Our transcriptomic analysis expanded the repertoire of nitrate-responsive genes/processes in rice and revealed their phenotypic association with root/shoot, stomata, tiller, panicle/flowering and yield, with agronomic implications for nitrogen use efficiency.
Abstract
Nitrogen use efficiency (NUE) is a multigenic quantitative trait, involving many N-responsive genes/processes that are yet to be fully characterized. Microarray analysis of early nitrate response in excised leaves of japonica rice revealed 6688 differentially expressed genes (DEGs), including 2640 hitherto unreported across multiple functional categories. They include transporters, enzymes involved in primary/secondary metabolism, transcription factors (TFs), EF-hand containing calcium binding proteins, hormone metabolism/signaling and methytransferases. Some DEGs belonged to hitherto unreported processes viz. alcohol, lipid and trehalose metabolism, mitochondrial membrane organization, protein targeting and stomatal opening. 1158 DEGs were associated with growth physiology and grain yield or phenotypic traits for NUE. We identified seven DEGs for shoot apical meristem, 66 for leaf/culm/root, 31 for tiller, 70 for heading date/inflorescence/spikelet/panicle, 144 for seed and 78 for yield. RT-qPCR validated nitrate regulation of 31 DEGs belonging to various important functional categories/traits. Physiological validation of N-dose responsive changes in plant development revealed that relative to 1.5 mM, 15 mM nitrate significantly increased stomatal density, stomatal conductance and transpiration rate. Further, root/shoot growth, number of tillers and grain yield declined and panicle emergence/heading date delayed, despite increased photosynthetic rate. We report the binding sites of diverse classes of TFs such as WRKY, MYB, HMG etc., in the 1 kb up-stream regions of 6676 nitrate-responsive DEGs indicating their role in regulating nitrate response/NUE. Together, these findings expand the repertoire of genes and processes involved in genomewide nitrate response in rice and reveal their physiological, phenotypic and agronomic implications for NUE.








Similar content being viewed by others
Data availability
Our raw microarray data that support the findings of this study have been deposited in NCBI-GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62164) with the accession number GSE62164 (GSM1520726, GSM1520727, GSM1520730, GSM1520731, GSM1520734 and GSM1520735). Additional transcriptome datasets were obtained from either the published supplementary materials and/or their authors cited in the article. All other datasets pertaining to the analyses are included in the supplementary information files.
Abbreviations
- AH:
-
Arnon-Hoagland nutrient solution
- DEG:
-
Differentially expressed gene
- NUE:
-
Nitrogen use efficiency
- PPI:
-
Protein–protein interaction
- TF:
-
Transcription factor
References
Abrol YP, Adhya TK, Aneja VP, Raghuram N, Pathak H, Kulshrestha U, Sharma C, Singh B (2017) The Indian nitrogen assessment: Sources of reactive nitrogen, environmental and climate effects, management options, and policies. Elsevier, Cambridge, MA, USA
Agrawal GK, Abe K, Yamazaki M, Miyao A, Hirochika H (2005) Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol Biol 59(1):125–135
Beatty PH, Carroll RT, Shrawat AK, Guevara D, Good AG (2013) Physiological analysis of nitrogen-efficient rice overexpressing alanine aminotransferase under different N regimes. Botany 91(12):866–883
Bhowmick R (2019) Comprehensive analysis of universal stress proteins and their promoter sequences in rice. Int J Curr Microbiol App Sci 8(7):1279–1286
Bluemel M, Dally N, Jung C (2015) Flowering time regulation in crops—what did we learn from Arabidopsis? Curr Opin Biotechnol 32:121–129
Cai H, Lu Y, Xie W, Zhu T, Lian X (2012) Transcriptome response to nitrogen starvation in rice. J Biosci 37(4):731–747
Chakraborty N, Raghuram N (2011) Nitrate sensing and signaling in genomewide plant N response. In: Jain V, Anandakumar P (eds) Nitrogen use efficiency in plants. New India Publishing Agency, New Delhi, pp 45–62
Chandran AKN, Priatama RA, Kumar V, Xuan Y, Je BI, Kim CM, Jung K-H, Han C-D (2016) Genome-wide transcriptome analysis of expression in rice seedling roots in response to supplemental nitrogen. J Plant Physiol 200:62–75
Coneva V, Simopoulos C, Casaretto JA, El-kereamy A, Guevara DR, Cohn J, Zhu T, Guo L, Alexander DC, Bi Y-M (2014) Metabolic and co-expression network-based analyses associated with nitrate response in rice. BMC Genomics 15(1):1056
Guo Y, Wu Q, Xie Z, Yu B, Zeng R, Min Q, Huang J (2020) OsFPFL4 is involved in the root and flower development by affecting auxin levels and ROS accumulation in rice (Oryza sativa). Rice 13(1):2. https://doi.org/10.1186/s12284-019-0364-0
He Y, Zhou J, Shan L, Meng X (2018) Plant cell surface receptor-mediated signaling–a common theme amid diversity. J Cell Sci 131(2):jcs209353. https://doi.org/10.1242/jcs.209353
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil, 2nd edn. Circular California agricultural experiment station, p 347
Hong W-J, Chandran AKN, Jeon J-S, Jung K-H (2017) Construction and application of functional gene modules to regulatory pathways in rice. J Plant Biol 60(4):358–379
Hooper CM, Castleden IR, Aryamanesh N, Jacoby RP, Millar AH (2016) Finding the subcellular location of barley, wheat, rice and maize proteins: the compendium of crop proteins with annotated locations (cropPAL). Plant Cell Physiol 57(1):e9–e9
Huang S, Liang Z, Chen S, Sun H, Fan X, Wang C, Xu G, Zhang Y (2019) A transcription factor, OsMADS57, regulates long-distance nitrate transport and root elongation. Plant Physiol 180(2):882–895
Jin X, Lv Z, Gao J, Zhang R, Zheng T, Yin P, Li D, Peng L, Cao X, Qin Y, Persson S, Zheng B, Chen P (2019) AtTrm5a catalyses 1-methylguanosine and 1-methylinosine formation on tRNAs and is important for vegetative and reproductive growth in Arabidopsis thaliana. Nucl Acids Res 47(2):883–898
Kamiya N, Itoh JI, Morikami A, Nagato Y, Matsuoka M (2003) The SCARECROW gene’s role in asymmetric cell divisions in rice plants. Plant J 36(1):45–54
Kronzucker H, Glass A, Siddiqi M, Kirk G (2000) Comparative kinetic analysis of ammonium and nitrate acquisition by tropical lowland rice: implications for rice cultivation and yield potential. New Phytol 145(3):471–476
Krouk G (2016) Hormones and nitrate: a two-way connection. Plant Mol Biol 91(6):599–606
Kumari S, Sharma N, Raghuram N (2021) Meta-analysis of yield-related and N-responsive genes reveals chromosomal hotspot, key processes and candidate genes for nitrogen use efficiency (NUE) in rice. Front Plant Sci 12:1006
Lacuesta M, Saiz-Fernández I, Podlešáková K, Miranda-Apodaca J, Novák O, Doležal K, De Diego N (2018) The trans and cis zeatin isomers play different roles in regulating growth inhibition induced by high nitrate concentrations in maize. Plant Growth Regul 85(2):199–209
Lee SK, Jeon JS, Boernke F, Voll L, Cho JI, Goh CH, Jeong SW, Park YI, Kim SJ, Choi SB (2008) Loss of cytosolic fructose-1,6-bisphosphatase limits photosynthetic sucrose synthesis and causes severe growth retardations in rice (Oryza sativa). Plant Cell Environ 31(12):1851–1863
Li H, Hu B, Chu C (2017) Nitrogen use efficiency in crops: lessons from Arabidopsis and rice. J Exp Bot 68(10):2477–2488
Li Y, Xiao J, Chen L, Huang X, Cheng Z, Han B, Zhang Q, Wu C (2018) Rice functional genomics research: past decade and future. Mol Plant 11(3):359–380
Li C, Ma F, Jiao R, Chen C, Wang Q, Xiao F, Sun C, Deng X, Dong C, Wang P (2019) Mutation in Mg-protoporphyrin IX monomethyl ester cyclase causes yellow and spotted leaf phenotype in rice. Plant Mol Biol Rep 37(4):253–264
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148:350–382
Lim J, Moon Y-H, An G, Jang SK (2000) Two rice MADS domain proteins interact with OsMADS1. Plant Mol Biol 44(4):513–527
Liu C, Xue Z, Tang D, Shen Y, Shi W, Ren L, Du G, Li Y, Cheng Z (2018) Ornithine δ-aminotransferase is critical for floret development and seed setting through mediating nitrogen reutilization in rice. Plant J 96(4):842–854
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4):402–408
Ma P, Liu J, Yang X, Ma R (2013) Genome-wide identification of the maize calcium-dependent protein kinase gene family. Appl Biochem Biotechnol 169(7):2111–2125
McLeay RC, Bailey TL (2010) Motif enrichment analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics 11(1):1–11
Meyer RC, Gryczka C, Neitsch C, Müller M, Bräutigam A, Schlereth A, Schön H, Weigelt-Fischer K, Altmann T (2019) Genetic diversity for nitrogen use efficiency in Arabidopsis thaliana. Planta 250(1):41–57. https://doi.org/10.1007/s00425-019-03140-3
Midorikawa K, Kuroda M, Terauchi K, Hoshi M, Ikenaga S, Ishimaru Y, Abe K, Asakura T (2014) Additional nitrogen fertilization at heading time of rice down-regulates cellulose synthesis in seed endosperm. PLoS One 9(6):e98738
Misyura M, Guevara D, Subedi S, Hudson D, McNicholas PD, Colasanti J, Rothstein SJ (2014) Nitrogen limitation and high density responses in rice suggest a role for ethylene under high density stress. BMC Genomics 15(1):681. https://doi.org/10.1186/1471-2164-15-681
Móring A, Hooda S, Raghuram N, Adhya TK, Ahmad A, Bandyopadhyay SK, Barsby T, Beig G, Bentley A, Bhatia A (2021) Nitrogen challenges and opportunities for agricultural and environmental science in India. Front Sustain Food Syst 5:505347
Obertello M, Shrivastava S, Katari MS, Coruzzi GM (2015) Cross-species network analysis uncovers conserved nitrogen-regulated network modules in rice. Plant Physiol 168(4):1830–1843
Pathak RR, Ahmad A, Lochab S, Raghuram N (2008) Molecular physiology of plant nitrogen use efficiency and biotechnological options for its enhancement. Curr Sci 94:1394–1403
Pathak RR, Jangam AP, Malik A, Sharma N, Jaiswal DK, Raghuram N (2020) Transcriptomic and network analyses reveal distinct nitrate responses in light and dark in rice leaves (Oryza sativa Indica var. Panvel1). Sci Rep 10(1):1–17
Paul P, Awasthi A, Rai AK, Gupta SK, Prasad R, Sharma T, Dhaliwal H (2012) Reduced tillering in Basmati rice T-DNA insertional mutant OsTEF1 associates with differential expression of stress related genes and transcription factors. Func Integr Genomics 12(2):291–304
Qi G-N, Yao F-Y, Ren H-M, Sun S-J, Tan Y-Q, Zhang Z-C, Qiu B-S, Wang Y-F (2018) The S-type anion channel ZmSLAC1 plays essential roles in stomatal closure by mediating nitrate efflux in maize. Plant Cell Physiol 59(3):614–623
Qin H, Li Y, Huang R (2020) Advances and challenges in the breeding of salt-tolerant rice. Int J Mol Sci 21(21):8385
Qin H, Wang J, Chen X, Wang F, Peng P, Zhou Y, Miao Y, Zhang Y, Gao Y, Qi Y (2019) Rice OsDOF 15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol 223(2):798–813
Raghuram N, Sutton MA, Jeffery R, Ramachandran R, Adhya TK (2021) From South Asia to the world: embracing the challenge of global sustainable nitrogen management. One Earth 4(1):22–27
Raghuram N, Sharma N (2019) Improving crop nitrogen use efficiency. In: Moo-Young M (ed) Comprehensive biotechnology, vol 4. Elsevier, pp 211–220. https://doi.org/10.1016/B978-0-444-64046-8.00222-6
Ramanathan V, Rahman H, Subramanian S, Nallathambi J, Kaliyaperumal A, Manickam S, Ranganathan C, Muthurajan R (2018) OsARD4 encoding an acireductone dioxygenase improves root architecture in rice by promoting development of secondary roots. Sci Rep 8(1):15713. https://doi.org/10.1038/s41598-018-34053-y
Reddy MM, Ulaganathan K (2015) RNA-Seq analysis of urea nutrition responsive transcriptome of Oryza sativa elite indica cultivar RP Bio 226. Genom Data 6:112–113
Sawaki N, Tsujimoto R, Shigyo M, Konishi M, Toki S, Fujiwara T, Yanagisawa S (2013) A nitrate-inducible GARP family gene encodes an auto-repressible transcriptional repressor in rice. Plant Cell Physiol 54(4):506–517
Séré D, Martin A (2020) Epigenetic regulation: another layer in plant nutrition. Plant Signal Behav 15(1):1686236
Sharma N, Sinha VB, Gupta N, Rajpal S, Kuchi S, Sitaramam V, Parsad R, Raghuram N (2018) Phenotyping for nitrogen use efficiency: rice genotypes differ in N-responsive germination, oxygen consumption, seed urease activities, root growth, crop duration, and yield at low N. Front Plant Sci 9:1452
Sharma N, Sinha VB, Prem Kumar NA, Subrahmanyam D, Neeraja C, Kuchi S, Jha A, Parsad R, Sitaramam V, Raghuram N (2021) Nitrogen use efficiency phenotype and associated genes: roles of germination, flowering, root/shoot length and biomass. Front Plant Sci 11:2329
Shin S-Y, Jeong JS, Lim JY, Kim T, Park JH, Kim J-K, Shin C (2018) Transcriptomic analyses of rice (Oryza sativa) genes and non-coding RNAs under nitrogen starvation using multiple omics technologies. BMC Genomics 19(1):532. https://doi.org/10.1186/s12864-018-4897-1
Souza LA, Tavares R (2021) Nitrogen and stem development: a puzzle still to be solved. Front Plant Sci 12:181
Sun L, Di D, Li G, Kronzucker HJ, Shi W (2017) Spatio-temporal dynamics in global rice gene expression (Oryza sativa L.) in response to high ammonium stress. J Plant Physiol 212:94–104
Sun Y, Wang M, Mur LAJ, Shen Q, Guo S (2020) Unravelling the roles of nitrogen nutrition in plant disease defences. Int J Mol Sci 21(2):572
Sutton MA, Bleeker A, Howard C, Erisman J, Abrol Y, Bekunda M, Datta A, Davidson E, De Vries W, Oenema O (2013) Our nutrient world. The challenge to produce more food & energy with less pollution. Centre for Ecology & Hydrology, https://edepot.wur.nl/249094. Accessed 3 May 2021
Sutton M, Raghuram N, Adhya TK, Baron J, Cox C, de Vries W, Hicks K, Howard C, Ju X, Kanter D (2019) The nitrogen fix: from nitrogen cycle pollution to nitrogen circular economy. Frontiers 2018/19: Emerging Issues of Environmental Concern. United Nations Environment Programme, Nairobi
Takehisa H, Sato Y, Antonio B, Nagamura Y (2015) Coexpression network analysis of macronutrient deficiency response genes in rice. Rice 8(1):24. https://doi.org/10.1186/s12284-015-0059-0
Takehisa H, Sato Y (2019) Transcriptome monitoring visualizes growth stage-dependent nutrient status dynamics in rice under field conditions. Plant J 97(6):1048–1060
Tsuji H, Taoka K-i, Shimamoto K (2011) Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr Opin Plant Biol 14(1):45–52
Udvardi M, Below FE, Castellano MJ, Eagle A, Giller KE, Ladha JK, Liu X, Mcclellan Maaz T, Nova-Franco B, Raghuram N (2021) A research road map for responsible use of agricultural nitrogen. Front Sustain Food Syst 5:165
Umate P, Tuteja R, Tuteja N (2010) Genome-wide analysis of helicase gene family from rice and Arabidopsis: a comparison with yeast and human. Plant Mol Biol 73(4–5):449–465
Vidal EA, Alvarez JM, Araus V, Riveras E, Brooks MD, Krouk G, Ruffel S, Lejay L, Crawford NM, Coruzzi GM (2020) Nitrate in 2020: thirty years from transport to signaling networks. Plant Cell 32(7):2094–2119
Wang R, Tischner R, Gutiérrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol 136(1):2512–2522
Wang Y-Y, Cheng Y-H, Chen K-E, Tsay Y-F (2018) Nitrate transport, signaling, and use efficiency. Annu Rev Plant Biol 69:85–122
Wang R, Qian J, Fang Z, Tang J (2020) Transcriptomic and physiological analyses of rice seedlings under different nitrogen supplies provide insight into the regulation involved in axillary bud outgrowth. BMC Plant Biol 20:197. https://doi.org/10.1186/s12870-020-02409-0
Wang X, Li Y, Fang G, Zhao Q, Zeng Q, Li X, Gong H, Li Y (2014) Nitrite promotes the growth and decreases the lignin content of indica rice calli: a comprehensive transcriptome analysis of nitrite-responsive genes during in vitro culture of rice. PLoS One 9(4):e95105
Wang D, Qin B, Li X, Tang D, Ye Z, Cheng Z, Xue Y (2016) Nucleolar DEAD-box RNA helicase TOGR1 regulates thermotolerant growth as a pre-rRNA chaperone in rice. PLoS Genet 12(2):e1005844
Wei H, Wang X, Xu H, Wang L (2020) Molecular basis of heading date control in rice. aBIOTECH 1:219–232
Wu K, Wang S, Song W, Zhang J, Wang Y et al (2020) Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 367(6478):eaaz2046. https://doi.org/10.1126/science.aaz2046
Xin W, Zhang L, Zhang W, Gao J, Yi J, Zhen X, Li Z, Zhao Y, Peng C, Zhao C (2019) An integrated analysis of the rice transcriptome and metabolome reveals differential regulation of carbon and nitrogen metabolism in response to nitrogen availability. Inter J Mol Sci 20(9):2349
Yang SY, Hao DL, Song ZZ, Yang GZ, Wang L, Su YH (2015a) RNA-Seq analysis of differentially expressed genes in rice under varied nitrogen supplies. Gene 555(2):305–317
Yang W, Yoon J, Choi H, Fan Y, Chen R, An G (2015b) Transcriptome analysis of nitrogen-starvation-responsive genes in rice. BMC Plant Biol 15:31. https://doi.org/10.1186/s12870-015-0425-5
Yang HC, Kan CC, Hung TH, Hsieh PH, Wang SY, Hsieh WY, Hsieh MH (2017) Identification of early ammonium nitrate-responsive genes in rice roots. Sci Rep 7:16885. https://doi.org/10.1038/s41598-017-17173-9
Yang S, Hao D, Jin M, Li Y, Liu Z, Huang Y, Chen T, Su Y (2020) Internal ammonium excess induces ROS-mediated reactions and causes carbon scarcity in rice. BMC Plant Biol 20:143. https://doi.org/10.1186/s12870-020-02363-x
Yu C, Liu Y, Zhang A, Su S, Yan A, Huang L, Ali I, Liu Y, Forde BG, Gan Y (2015) MADS-box transcription factor OsMADS25 regulates root development through affection of nitrate accumulation in rice. PLoS One 10(8):e0135196
Zhao H, Frank T, Tan Y, Zhou C, Jabnoune M, Arpat AB, Cui H, Huang J, He Z, Poirier Y (2016) Disruption of OsSULTR 3; 3 reduces phytate and phosphorus concentrations and alters the metabolite profile in rice grains. New Phytol 211(3):926–939
Zhuang L, Ge Y, Wang J, Yu J, Yang Z, Huang B (2019) Gibberellic acid inhibition of tillering in tall fescue involving crosstalks with cytokinins and transcriptional regulation of genes controlling axillary bud outgrowth. Plant Sci 287:110168
Acknowledgements
This work was supported by research grants to NR from ICAR (F. No. 2-2(60)/10-11/NICRA), Department of Biotechnology (DBT) [BT/IN/UK-VNC/44/NR/2015-16], UKRI GCRF South Asian Nitrogen Hub (SANH) [NE/S009019/1] GGSIPU [GGSIPU/DRC/Ph.D/Adm/2016/1549], [GGSIPU/DRC/FRGS/2018/22] and [GGSIPU/DRC/FRGS/2019/1553/24]. Fellowships were paid to VKM from DBT (DBT/JRF/14/AL/445) and GGSIPU (STRF:GGSIPU/DRC/2020/2049), APJ from CSIR (09/806(013)2008-EMR-I) and NC from UKRI GCRF-SANH [NE/S009019/1]. We thank Prof. T. Kumamaru from Kyushu University, Japan, for providing the rice seeds and the Regional Centre for Biotechnology (RCB), Faridabad for help with the scanning electron microscopy. We thank Dr. Dinesh Kumar Jaiswal for his assistance in early data analysis and initial draft of the manuscript. We also thank to Prof. Chanseok Shin from Seoul National University, Korea and Dr. Rumei Chen from Chinese Academy of Agricultural Sciences, China for providing their RNA-seq data for our meta-analyis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mandal, V.K., Jangam, A.P., Chakraborty, N. et al. Nitrate-responsive transcriptome analysis reveals additional genes/processes and associated traits viz. height, tillering, heading date, stomatal density and yield in japonica rice. Planta 255, 42 (2022). https://doi.org/10.1007/s00425-021-03816-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-021-03816-9