Abstract
Post-transcriptional regulation of gene expression plays a critical role in almost all cellular processes. Regulation occurs mostly by RNA-binding proteins (RBPs) that recognise RNA elements and form ribonucleoproteins (RNPs) to control RNA metabolism from synthesis to decay. Recently, the repertoire of RBPs was significantly expanded owing to methodological advances such as RNA interactome capture. The newly identified RNA binders are involved in diverse biological processes and belong to a broad spectrum of protein families, many of them exhibiting enzymatic activities. This suggests the existence of an extensive crosstalk between RNA biology and other, in principle unrelated, cell functions such as intermediary metabolism. Unexpectedly, hundreds of new RBPs do not contain identifiable RNA-binding domains (RBDs), raising the question of how they interact with RNA. Despite the many functions that have been attributed to RNA, our understanding of RNPs is still mostly governed by a rather protein-centric view, leading to the idea that proteins have evolved to bind to and regulate RNA and not vice versa. However, RNPs formed by an RNA-driven interaction mechanism (RNA-determined RNPs) are abundant and offer an alternative explanation for the surprising lack of classical RBDs in many RNA-interacting proteins. Moreover, RNAs can act as scaffolds to orchestrate and organise protein networks and directly control their activity, suggesting that nucleic acids might play an important regulatory role in many cellular processes, including metabolism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The landscape of RNA-binding proteins
RNA assembles with proteins forming dynamic complexes named ribonucleoproteins (RNPs). The sequence of the transcript, its processing and the activity of the available RNA-binding proteins (RBPs) determine the composition of the RNP [42, 82], establishing an additional layer of information (the RNP code) that determines the fate of the RNA, shaping transcriptome and proteome. RNPs are not static, and their remodelling allows adjustments in gene expression under conditions that require adaptive changes. Dysfunction of RBPs is often linked to disease [19, 26, 28, 75, 88], which reflects the relevance of protein-RNA interactions in cellular homeostasis.
Given the complexity of RNA metabolism, it was estimated that RBPs comprise 3–11 % of the proteome of bacteria, archaea and eukaryotes and they are significantly more conserved across evolution than proteins lacking RNA-binding activity [3, 40]. Most of the RBPs discovered over the last three decades match the classical view of RBP architecture with a modular combination of well-characterised RNA-binding domains (RBDs) such as the RNA recognition motif (RRM), the K homology domain (KH) and the DEAD box helicase domain [76]. Individual RBDs recognise short stretches of RNA (approx. 2–10 nucleotides in length) with often low affinity. RBPs usually build their affinity and specificity for RNA on the cooperative function of multiple classical RBDs [49, 76], as exemplarily illustrated by the four RRMs that work together in nucleolin (NCL) or the poly(A)-binding protein (PABP).
A number of studies in the last two decades reported RNA-binding activities in proteins lacking classical RBDs, suggesting that the scope of RBPs was initially underestimated. These newly discovered RNA binders contain protein domains with dual function (e.g. enzymatic and RNA-binding activities) [114], folds of unknown function [54] and protein regions lacking a defined tertiary structure in the unbound state [95]. Given the growing evidence of unorthodox RNA-binding activities within proteins previously unrelated to RNA biology [24, 50, 86], system-wide approaches were developed aiming at determining the complete repertoire of RBPs.
In silico approaches were successfully used to identify proteins harbouring classical RBDs and highly homologous protein domains that may likely bind RNA as well [3, 40, 65]. However, the capacity of these methods to discover RNA-binding architectures that lack similarities with classic RBDs was very limited. To circumvent this, different in vitro and in vivo approaches for comprehensive identification of RBPs were developed. In two parallel studies, Scherrer et al. and Tsvetanova et al. used protein arrays to identify RBPs in Saccharomyces cerevisiae. A significant fraction of the yeast proteome (∼4000 proteins) was purified and immobilised on a membrane. The resulting array was subsequently incubated with a mixture of fluorophore-labelled RNAs, and fluorescence retained at each protein spot was used as readout for RNA binding. Using this method, these studies identified 180 [103] and 42 [111] proteins, respectively, as RBPs. Surprisingly, many of the identified proteins were not related to RNA biology, including dozens of metabolic enzymes such as oxidoreductases and proteins involved in lipid metabolism, calling for further in vivo validation of their RNA-binding activities.
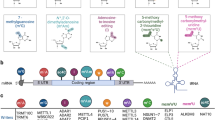
More recently, two independent works described a new method for the unbiased and comprehensive identification of RBPs from living cells, which is referred to as RNA interactome capture [8, 18]. In brief, native protein-RNA interactions are covalently immobilised by applying ultraviolet (UV) light to cultured cells [45, 52, 92]. UV irradiation induces short-lived free radicals at the nucleotide base that can attack amino acids in close proximity. Because proteins do not efficiently absorb UV light at these wavelengths, protein-protein cross-linking is not detectable [18, 107]. After irradiation, a stringent purification of polyadenylated (poly(A)) RNA is performed under denaturing conditions, followed by identification of co-purified, cross-linked proteins by quantitative mass spectrometry. The initial RNA interactome studies lead to the identification of 1106 RBPs in HeLa [18] and HEK293 [8] cells, with an extensive overlap between the two datasets (545 proteins). Both datasets do not only validate RNA binding of known RBPs, but they are also catalogued as novel RNA binders hundreds of proteins previously unrelated to RNA metabolism (561 proteins) [8, 18]. The newly identified RBPs belong to different protein families, participate in multiple biological processes, mediate distinct molecular functions and, surprisingly, in most of the cases, lack known RBDs (Fig. 1).
RNA interactome capture discovers many RNA-binding proteins that lack identifiable RNA-binding domains. RNA interactome capture from different human cell lines [8, 13, 18] identified a total of 1218 proteins as RNA binders, most of which do not contain an identifiable RBD (∼55 %). The remaining proteins harbour domains known to bind RNA, most commonly the RNA recognition motif (RRM, accounting for ∼13 % of the proteins), DEAD/DEAD box helicase domain (accounting for ∼4 %) and the K homology domain (KH, accounting for ∼3 %)
Subsequently, this methodology was extended to determine RNA interactomes in different cell lines and species. Kwon et al. added 283 novel RNA-binding proteins from mouse embryonic stem cells to the growing list of RBPs. Sixty-eight of these proteins are highly expressed in undifferentiated cells, suggesting a role in stem cell physiology [63]. Three independent studies identified 120 [81], 765 [78] and 678 [13] RNA binders in S. cerevisiae. Surprisingly, many enzymes from classical biochemical pathways, particularly from glycolysis, were found to moonlight as RNA-binding proteins in this unicellular organism. And finally, the interactome of an entire multicellular organism was derived from the worm Caenorhabditis elegans, identifying 594 poly(A) RNA-interacting proteins [78].
Comparison of the datasets revealed a conserved core of eukaryotic RNA-binding proteins, comprising ∼250 RBPs [13], most of which have known functions in RNA biology, e.g. in transcription, translation, RNA transport, degradation and/or modification. Surprisingly, around 9 % of the conserved RBPs also have reported enzymatic activities [13]. Presumably, datasets from additional species, including the most common eukaryotic model organisms (e.g. Drosophila melanogaster, Danio rerio, Arabidopsis thaliana) will become available soon, enabling further inter-species comparisons and even deeper insights into RNA biology.
What sequences do the (novel) RBPs bind?
With the world of RBPs rapidly expanding and with more and more RBDs being identified, the need arises for a rapid identification of their binding motifs and target RNAs to gain insight into RBP function.
Numerous experimental methods have been developed to study protein-RNA interactions. These ribonomic analyses range from the determination of in vitro binding specificities of recombinant RNA-binding proteins (or domains thereof) to the purification and subsequent analysis of native RNPs. Employing a panel of recombinant and purified RBPs in single and competitive binding reactions against a complex pool of RNA ligands in vast excess (RNAcompete) has recently yielded the binding specificities of 207 RBPs from various species. Not surprisingly, it also revealed that two proteins that share a high degree of identity between their RBDs are likely to have similar or even identical RNA sequence specificity [98]. Another powerful method involves immunopurification of native RBPs from cell extracts and analysis of bound RNAs either by hybridisation to microarrays (RIP-Chip) or high-throughput sequencing (RIP-Seq) [84]. As described above for interactome capture, in vivo UV cross-linking has been employed to stabilise the highly dynamic environment of ribonucleoproteins and to preserve protein-RNA interactions through stringent affinity purification protocols, followed by the subsequent analysis of co-purified RNA (cross-linking and immunoprecipitation (CLIP), CRAC and related methods) [5]. The ‘freezing’ of protein-RNA interactions by UV cross-linking in living cells (or entire organisms) ensures that only interactions are captured that occur under native, cellular conditions. This is particularly important, as concerns were raised, that protein-RNA interactions which do not necessarily reflect the in vivo situation can occur after cell lysis in the extract [80, 100, 101].
Common to the aforementioned methods is that they do not only test for binary interactions of one protein (or RBD) with a single RNA species (or short oligonucleotide) but rather produce global or transcriptome-wide binding profiles. However, in most cases, studies are limited to individual RNA-binding proteins (or their respective RBDs). Expanding the analyses to other RBPs has been hampered either by the complexity of the experimental procedures, the availability of antibodies or the requirement for recombinant protein production.
Structural analyses of protein-RNA complexes have resulted in the identification of interaction surfaces and amino acids of proteins that are directly involved in RNA binding. This has significantly advanced our understanding of the principles that underlie binding specificity and RNP formation [6, 17]. Moreover, structures that contain multiple RBDs in complex with their ligand have unveiled how protein domains cooperate to recognise longer, continuous stretches of RNA to increase both RNA affinity and specificity [49]. However, our current structural knowledge of protein-RNA complexes is still rather limited. While the protein data bank (as of January 2016) lists more than 116,000 protein (or protein domain) structures, only less than 1.7 % (approx. 1800) are in complex with RNA. Moreover, the vast majority of the structural information on RNPs is derived from X-ray crystallography that requires rigid folds, such as globular protein domains. Hence, intrinsically unfolded proteins that bind RNA (see below) are highly underrepresented.
Understanding RNA-binding specificity of proteins
Being able to predict the target RNA motif(s) based on the amino acid sequence of an RBD would greatly advance our understanding of RBPs and facilitate prediction of their cellular function(s), even if the proteins have not yet been extensively studied. However, the molecular basis of sequence-specific recognition of nucleic acids is well understood for only a subset of RNA-binding domains.
Pumilio and FBF homology (PUF) proteins employ a repetitive and modular scaffold for sequence-specific binding to RNA, each repeat recognising one nucleobase [116]. Breaking of this recognition code has allowed a rational design of custom proteins, tailored to recognise specific RNA sequences of interest, producing numerous genetically encoded tools to study RNA biology [117]. But despite the accumulating knowledge about RNP biology, understanding, predicting and engineering the specificity of RBDs other than the PUF domain remains a challenge. Hence, binding motif inference based on known specificities of closely related proteins still remains an approximation that requires experimental confirmation.
Moreover, RBPs do not necessarily only bind in a sequence-specific manner, recognising a stretch of specific nucleobases. The exon junction complex (EJC, its core comprising the proteins eukaryotic initiation factor (eIF)4AIII, Y14, Magoh and MLN51) has no apparent sequence specificity and is deposited ∼20 nucleotides upstream of exon-exon junctions by the splicing machinery. Its binding to the RNA is remarkably stable, allowing the EJC to stay associated with the RNA during nuclear export and requiring a dedicated disassembly factor in the cytoplasm [39, 64]. Other proteins recognise an RNA structure rather than a sequence or both. The protein Staufen 1 regulates messenger RNA (mRNA) localisation, stability and translation through binding to double-stranded RNA motifs of variable sequence. Analysis of its binding sites by CLIP has not only led to the identification of its RNA targets but also revealed thousands of RNA regions that form duplexes in vivo [99, 108]. Last but not least, a single protein (or protein complex) can recognise and bind to different RNA elements that are diverse in sequence and structure. This is exemplified by the RNA-binding protein She2p from S. cerevisiae that, together with its partner She3p, associates with different cis-acting RNA elements (so-called ‘zipcodes’) to mediate subcellular transport and localization of mRNAs [51, 87].
The complexity of protein-RNA interactions is probably best illustrated by the RNA recognition motif. Although being the most abundant and best-studied RBD, the RRM is also one of the most versatile. Despite a shared architecture, RRMs utilise different surfaces for RNA binding and display various different modes of interaction, thus making target prediction extremely difficult [6, 30]. Moreover, the RRM-type protein fold can also be employed for the interaction with proteins instead of RNA, giving rise to U2AF homology motifs (UHMs). These domains are highly similar to classical RRMs but lack some of the critical amino acids involved in RNA recognition, having instead evolved sequence characteristics that optimise the interaction with peptide ligands [60]. Interestingly, some RRMs can engage in both RNA and protein binding. This has been reported e.g. for the RRM domain of eIF3b, which associates with eIF3j or hepatitis C virus mRNA, presumably in a mutually exclusive manner [34, 93]. Another example is the RRM2 of the PABP that simultaneously interacts in a cooperative manner with the eIF4G and the poly(A) tail of the RNA to support translation initiation [102].
Taken together, this further complicates prediction of binding motifs and also highlights that in the cellular context, interactions between RBPs and their targets occur in a complex environment with many different binding partners available. Moreover, association with one ligand often impacts on interactions with other factors. Both mutually exclusive binding and highly synergistic binding have been reported for a number of RBPs. One example is the Drosophila RNA-binding protein Sex lethal (Sxl) which harbours two RRMs that bind U-rich sequences with high affinity. On the one hand, Sxl can compete with and evict other RNA-binding proteins or complexes such as U2AF, CstF64 and others [36, 37, 44], and on the other hand, it also acts as a nucleation factor to recruit proteins such as Upstream of N-ras (Unr) or Held-out wings (How) [1, 33, 44, 48]. This way, Sxl acts as a remodeler of RNPs, modulating their composition to control RNA fate.
Synergistic RNA binding of two proteins does not always necessitate a direct interaction between the two RBPs. Neither do antagonising factors always have to compete for binding to the same RNA element to exhibit a mutually exclusive binding behaviour. The RNA itself can relay the information of a binding event to a distal site through structural rearrangements. This can either improve accessibility or impede interaction with other factors. Exemplarily, the 3′ untranslated region (UTR) of vascular endothelial growth factor (VEGF) mRNA shows mutually exclusive and stimulus-dependent binding of two protein complexes, the HILDA (hypoxia-inducible hnRNP L-DRBP76-hnRNP A2/B1) complex and the interferon gamma (IFN-γ)-activated inhibitor of translation complex (GAIT). Binding of the HILDA complex results in a conformational change of the RNA that occludes the binding site of the GAIT complex. This creates a binary, molecular switch to adjust gene expression in response different stimuli [97, 124].
Despite the complexity of the interactions and experimental challenges to characterise RNPs, detailed insights into binding specificities and target RNAs have been obtained for numerous proteins [5]. This has revealed that in many cases, RBPs associate with mRNAs encoding functionally related proteins, forming the so-called ‘RNA operons’ [58]. Thus, a limited set of (or even individual) RBPs can control entire pathways through the coordinate regulation of functionally related transcripts, fine-tuning and adjusting gene expression to the cellular requirements.
In light of this, RNA interactome capture could well prove a treasure trove for RNA biologists. Experimentation can now be extended to proteins that previously had not been implicated in RNA biology but were identified as novel RNA binders by interactome capture, resulting in the discovery of novel RBDs. Exemplarily, interactome capture has contributed to the identification of the NHL domain (named after the NCL-1, HT2A and LIN-41 proteins) as a bona fide RBD. Several NHL domain-containing proteins were found to co-purify with RNA [8, 18, 63]. Subsequent biochemical and structural studies confirmed sequence-specific RNA-binding activity and discovered functions in post-transcriptional regulation of gene expression [63, 69–71].
Furthermore, identification of the RNA targets of newly discovered RBPs will provide insights into their cellular function and further advance our understanding of RNA metabolism. This is exemplified by the fas-activated serine/threonine kinase (FASTK) family. In humans, six FASTK protein family members are present and all of them co-purify with RNA in interactome profiling [18]. FASTK proteins harbour a RAP domain (RNA-binding domain abundant in apicomplexans) which exhibits a high degree of homology with the bacterial endonuclease-like fold and was predicted to bind RNA [18, 65]. Loss of the RAP domain in FASTKD2 is linked to mitochondrial cytochrome C oxidase-defective encephalomyopathy, a rare neurological disorder [41]. Recent work identified FASTKD2 as a component of mitochondrial RNA granules, implicating a function in mitochondrial RNA processing [4]. Finally, the identification of its target RNAs has paved the way for functional studies and established a link between FASTKD2 activity, mitochondrial dysfunction and human disease [96]. This example highlights how RNA interactome studies can guide experimentation in the discovery of unprecedented aspects of RNA biology.
In light of this, the newly identified RBPs and the growing number of novel RBDs that have been functionally validated by independent laboratories [13, 23, 38, 66, 88] predict a new age in RNA research with exciting discoveries to be made.
RNA binding through low-complexity protein regions
Besides the abundance of proteins without a recognisable, classical RBD, large-scale datasets of RNA binders harbour another surprising finding: the identified proteins are enriched in low-complexity regions mainly composed of the amino acids serine (S), proline (P), glycine (G), arginine (R), lysine (K) and tyrosine (Y) [18]. These amino acids do not combine randomly but rather form defined patterns: G often co-occurs with R or Y, generating RG or YG repeats that can appear multiple times within a given protein region giving rise to highly repetitive sequences. Both serine and proline have a high propensity to be in disordered regions [68], and it has been predicted that many of the low-complexity regions in RBDs are intrinsically disordered regions (IDRs) which natively lack stable three-dimensional structure [16, 18, 46].
Interestingly, also ribosomal proteins often contain long extensions enriched in the amino acids G, R and K (Fig. 2a). While being flexible and often disordered in solution, these protein tails are found to adopt specific conformations in the ribosome, reaching deep into the core of the RNP and forming extensive interactions with the ribosomal RNA (Fig. 2b) [14, 15, 61]. Here, the flexibility and small size of G allows tight packing against the RNA, while the basic amino acids R and K contribute to RNA folding by mediating electrostatic interactions and neutralising the negative charge of the RNA backbone [61]. Similarly, unstructured tails rich in basic amino acids have been reported to play a role in protein-DNA interaction. By projecting into the minor grove of the DNA double strand, they increase local affinity and promote hopping and linear diffusion along the DNA molecule [112, 113]. And finally, a low-complexity, repetitive motif, the arginine-glycine-rich RGG box motif that is found in numerous RBPs [110], was demonstrated to interact with RNA [29]. In the fragile X mental retardation protein (FMRP), an RGG box binds with high affinity to guanidine-rich RNA sequences. This interaction is mediated by a combination of shape complementarity facilitated by the flexible G linker and the electrostatic potential of the arginines [95].
Protein-RNA interaction through low-complexity, extended protein regions and protein-binding RNAs. a, b The S. cerevisiae ribosomal protein L23 (RPL23) contains a C-terminal domain that folds into an RRM-like topology, whereas the N-terminal, low-complexity region adopts an idiosyncratic, extended conformation (a). In the 60S ribosomal subunit, the lysine- and arginine-rich N-terminal domain of RPL23 participates in extensive interactions with the ribosomal 25S and 5.8S RNAs (b). RNAs are depicted in grey, neighbouring proteins RPL8 and RPL35 in light blue; in the structure based on Ben-Shem et al. [14] (PDB ID: 4V88), residues within 20A of the RPL23 protein are depicted. c HCV IRES bound to a 40S ribosomal subunit. The HCV IRES (blue) displays an elongated structure that binds the solvent side of the 40S ribosomal subunit. Interactions are formed mostly with ribosomal proteins (eS1/S3A, uS7/S5, eS7/S7, uS11/S14, eS25/S25, eS26/S26, eS27/S27 and eS28/S28, proteins depicted in yellow) and, only to a lesser extent, with the ribosomal 18S RNA (shown in grey). In the structure based on Yamamoto et al. [123] (PDB ID: 5FLX), individual panels represent different orientations (rotated by 90°)
In addition to directly interacting with nucleic acids, IDRs in RNA-binding proteins have been shown to contribute to assembly and formation of RBP ultrastructures such as P bodies, stress granules or nuage [16]. They can directly promote quick phase transitions, resulting in the formation of droplets—membrane-free, dynamic cellular subcompartments that harbour specific RNPs [53, 89]. Acting as assembly domains, IDRs are capable of recruiting other IDR-containing proteins to these granules, enabling the formation of specialised compartments by phase transition. This molecular aggregation results in the formation of hydrogels and amyloid-like fibres, and misregulation of aggregation has been linked to several neurological disorders [47, 56, 67, 120].
Comparison of RNA interactome datasets from different organisms has revealed an expansion in the number of short linear motifs in RBPs in higher eukaryotes compared to unicellular yeast, while the number of classical RBDs remains similar [13]. In sum, the occurrence of IDRs might explain the RNA-binding properties of ∼100 novel human RBPs that lack globular RBDs [18].
RNA-binding proteins and protein-binding RNAs
Surprisingly, interactome capture has also identified a number of proteins as RNA binders which do neither contain an identifiable globular RBD nor a predicted IDR. In human cell lines, more than half of the identified RBPs have no detectable RBD and have not been associated with functions in RNA metabolism before (Fig. 1) [8, 13, 18]. Similar results were obtained for rodents [63] and C. elegans [78], and an even larger fraction of proteins that lack identifiable RBDs were observed in yeast [13, 78, 81]. This raises an important question: are these proteins ‘false positives’ in interactome analyses, or do they contain novel (and yet unidentified) RBDs and/or bind RNA by non-conventional means?
Alike to proteins, also RNA can fold into intricate three-dimensional structures that serve various functions including catalytic activity, a classical example being the ribosome—a macromolecular and complex RNA machinery. Moreover, it has been demonstrated by systematic evolution of ligands by exponential enrichment (SELEX) that RNA sequences can be derived (so-called ‘RNA aptamers’) to selectively bind various different classes of small molecules [43]. Examples can also be found in nature: some bacterial mRNAs harbour sequences/structures with high affinity and specificity for various, structurally diverse metabolites. Ligand binding usually results in a conformational change of the RNA and in altered gene expression, forming a so-called ‘riboswitch’ that allows tailoring of protein production according to the nutritional status and cellular requirements [90].
Insofar, it is surprising that our understanding of RNPs is still mostly governed by a rather protein-centric view and the idea that proteins evolve to bind RNA and not vice versa. Often, it is difficult to differentiate between these two possibilities and there is clear evidence for co-evolution of proteins and RNA that interact to form an RNP. In fact, a continuous and densely populated spectrum exists, with one extreme being protein-determined RNPs (represented by many mRNPs) and the other one being RNA-determined RNPs like ribosomes [126]- the latter often considered relics, reminiscent of an RNA world in early evolution. In many RNA-determined RNPs such as ribosomes and other ribozymes, the enzymatic function is provided by the RNA, exposing it to much higher evolutionary pressure and often limiting the function of the bound proteins to being folding catalysts (RNA chaperones).
It is tempting to speculate that in RNA interactome datasets, the abundance of proteins that lack identifiable RBDs might, to some extent, reflect the capture of RNA-determined RNPs. In this case, RNAs have evolved to interact with proteins which hence do not require canonical RBDs.
An example of RNA-determined RNPs can be observed in a number of viral RNAs. Complex RNA secondary structures which promote viral translation can be found in the 5′ untranslated region of many different viruses, among them are picornaviruses, hepatitis C virus, herpes simplex virus and others. These so-called internal ribosome entry sites (IRESs) allow non-canonical and cap-independent translation initiation by functionally replacing some (or even all) eukaryotic translation initiation factors (eIFs), which are otherwise needed for normal, cap-driven translation [32]. Cap-dependent translation initiation is complex and requires the step-wise and hierarchical assembly of an RNP that then serves to recruit a small ribosomal subunit. By directly binding and stably recruiting specific eIFs (or even entire ribosomal subunits), viral IRESs can bypass some of the initial assembly steps, essentially becoming independent of initiation factors that act early. Exemplarily, the IRES of encephalomyocarditis virus binds directly to eIF4G, bypassing the requirement for eIF4E [72, 94], and the hepatitis C virus IRES interacts directly with the eIF3 complex and small ribosomal subunits (Fig. 2c) [74, 105, 109]. The binding of HCV IRES to eIF3 requires a certain geometry of the RNA and involves specific bases at critical positions [59], and structural studies have revealed extensive interactions between the IRES and eIF3, covering a broad surface area [104]. Moreover, an association of the HCV IRES with small ribosomal subunits employs mostly interactions with ribosomal proteins and only, to a lesser extent, interactions with the 18S ribosomal RNA (Fig. 2c) [123]. Even though classical RNA-binding domains of protein interaction partners appear to be involved to some extent, the interactions with the IRES involve surface areas of the proteins that differ from the ones that are usually employed for canonical interactions [93]. In sum, this supports the notion that the viral RNA evolved to specifically bind cellular proteins to support viral translation. IRES activity has also been reported for some cellular RNAs; however, factor requirements and interactions are not nearly as well characterised as for their viral counterparts [35, 121].
What appears to be a common theme to aptamers and IRES elements is that ligand/protein binding requires (extensive) RNA structure elements. While in rare cases promoting internal translation initiation, extensive RNA structures in 5′ UTRs or even coding regions of mRNAs are otherwise inhibitory to translation. Hence, mRNAs whose major role is to serve as templates for protein synthesis are less likely to contain such intricate structural elements that function in protein binding. However, for non-coding RNAs, these constraints do not apply and one might envision that RNA aptamers that bind proteins might be more common in this class of RNAs. In fact, many of the well-studied non-coding RNAs form extensive secondary structures to associate with proteins. This includes several classes of abundant RNAs such as small nuclear RNAs (snRNAs) involved in splicing [122], small nucleolar RNAs (snoRNAs) that direct RNA modification [119], the RNA moiety of the signal recognition particle (SRP RNA or 7SL RNA) critical for membrane protein insertion and protein secretion [73], RNase P RNA essential for transfer RNA (tRNA) processing [83] and telomerase RNA involved in chromosome maintenance [127].
Functional and structural roles of RNA
The function of RNA extends well beyond the mere coding for peptides. Exemplarily, RNA is not only the major structural component of ribosomes but also contributes catalytic activity and delivers amino acids for protein synthesis. Moreover, RNAs play integral roles in RNA processing and modification, are involved in sensing of metabolites and act as scaffolds to organise larger complexes. Several classes of small non-coding RNAs provide specificity for proteins, acting as guides. In eukaryotes, microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-associated RNAs (piRNAs) assemble with proteins of the argonaute clade and recruit them to their nucleic acid targets by (partial) sequence complementarity, eliciting gene regulatory pathways [79]. Similarly, snoRNAs and the closely related small Cajal body-specific RNAs (scaRNAs) direct RNA modification, determining targets through base paring interactions [119]. In prokaryotes, clustered regularly interspaced short palindromic repeat (CRISPR)-Cas immune systems rely on RNAs to identify foreign nucleic acids, targeting them for degradation [77]. Here, CRISPR RNAs (crRNAs) and trans-encoded crRNAs (tracrRNAs) serve as guides for Cas nucleases to direct DNA cleavage [55]. Harnessing the power of Cas nucleases and their RNA guides has revolutionised genome editing in many organisms [22, 55, 77, 106].
In telomerase, the RNA moiety does not only serve as a template for synthesis of telomeric repeats by the telomerase reverse transcriptase (TERT) but also provides a flexible scaffold to assemble the entire RNP, providing binding platforms for the Ku heterodimer, the LSm heteroheptameric protein complex and TERT [126].
Recently, long non-coding RNAs (lncRNAs), a heterogeneous and functionally diverse group of transcripts in eukaryotes, have gained widespread attention. They are polyadenylated, often contain extensive secondary structure and elicit different functions, serving e.g. as scaffolds, decoys and/or guides [62, 115]. Exemplarily, the formation of paraspeckles, a nuclear compartment rich in factors with roles in RNA processing, depends on the lncRNA nuclear-enriched autosomal transcript 1 (NEAT1). The RNA plays an architectural role, interacting with and nucleating several proteins such as paraspeckle protein 1 (PSP1); splicing factor, proline- and glutamine-rich (SFPQ) protein; and p54nrb/NONO (non-POU-domain-containing octamer-binding protein). Knockdown of the RNA results in a loss of paraspeckles and redistribution of proteins [25], reflecting the importance of the RNA for the spatial organisation of this compartment.
RNA as a regulator of protein activity
The structural role of RNA in the organisation of RNPs, where it e.g. serves as a (flexible) scaffold, is well documented, and its function as guide for proteins, directing them to their sites/sequences of action, is also well established. But can RNAs also regulate the activity of their bound protein partners?
Some of the best-documented examples, highlighting the function of RNAs as regulators of protein activity, are found in innate immunity. Toll-like receptors involved in the pattern recognition of pathogens are activated by double-stranded RNA (TLR-3), single-stranded RNA (TLR-7/8) and bacterial ribosomal RNA (TLR-13) [57, 91, 125]. The activity of the cytoplasmic retinoic acid-inducible gene I (RIG-I)-like receptors, nucleotide oligomerization domain (NOD)-like receptors and interferon-induced proteins with tetratricopeptide repeats (IFITS) depends on recognition of RNAs with 5′-triphosphate structures [2, 9]. And finally, protein kinase R (PKR) is activated by long double-stranded RNA (dsRNA) which is often generated during viral replication. dsRNA can trigger PKR dimerisation resulting in autophosphorylation and activation [7, 27]. Among the PKR targets is the eIF2α that plays critical role in translation initiation, escorting the initiator tRNA to the small ribosomal subunit. Phosphorylation of eIF2α by PKR traps it in an inactive state [31], impairing cellular translation and viral replication.
But RNA regulation of protein activity is neither restricted to innate immunity nor eukaryotes. Bacterial 6S RNA is a ∼200-nt-long, non-coding RNA which inhibits transcription of housekeeping genes in Escherichia coli and Bacillus subtilis. It adopts a rod-shaped secondary structure with a flexible central region, thereby mimicking an open promoter to bind and inhibit the bacterial RNA polymerase [118]. 6S RNA is one of the most abundant RNA species in bacterial cells during stationary growth phase and was identified in all branches of the bacterial kingdom [11]. While the sequence of this ncRNA varies widely between species, its secondary structure is predicted to be highly conserved among prokaryotes as well as its binding to RNA polymerase and its inhibitory activity [12]. Other RNAs that directly interact with the transcription machinery to regulate gene expression have also been identified in eukaryotic organisms and viruses [10].
In light of RNA being a regulator of protein function, it is intriguing that many of the proteins that are found to associate with RNA are reported to exhibit enzymatic activities. Dozens of enzymes from intermediary metabolism associate with RNA, including almost all glycolytic enzymes [13, 21, 85, 86]. A particularly interesting example is glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a key enzyme in glycolysis that has recently been reported to interact with the 3′ UTR of the IFN-γ mRNA, inhibiting its translation [21]. The RNA-binding activity of GAPDH depends on the glycolytic activity of the cell: upon activation, the metabolism of T lymphocytes shifts from oxidative phosphorylation to aerobic glycolysis, preventing the association of GAPDH with RNA and allowing IFN-γ translation [21]. Similarly, cellular iron metabolism controls the function of the iron regulatory protein (IRP) 1. In iron-deficient cells, IRP1 binds to and regulates translation and stability of various RNAs; however, in the presence of iron, it assembles an iron sulfur cluster (4Fe-4S) and functions as a cytoplasmic aconitase [20, 85, 114]. GAPDH and IRP1 present two interesting examples of mutually exclusive regulation: RNA binding is incompatible with enzymatic activity and protein function switches from metabolism to post-transcriptional regulation of gene expression.
Taken together, RNA is more than a mere bystander molecule that is being regulated by proteins. Various RNAs can play active roles in the organisation of ribonucleoproteins, sometimes even shaping entire subcellular compartments. Moreover, RNAs can control the function and activity of their bound protein partners, adjusting cell physiology to changing cellular requirements. In sum, this has fuelled the hypothesis that RNA may orchestrate enzymatic activities by bringing together all the enzymes that are part of a given metabolic pathway into large protein assemblies, forming metabolons with superior metabolic performance [20].
Scaffolding, protein binding and regulation of enzymatic activity have, however, only been described for a limited set of RNAs. The large number of unconventional RNA-binding proteins that were recently discovered might be a hint that many more examples of functionally active RNAs are waiting to be discovered. Characterisation and functional studies of RNPs that contain novel RNA-binding proteins with enzymatic activities will pave the way for a better understanding of how RNA biology integrates with other, in principle unrelated, cell functions such as intermediary metabolism.
References
Abaza I, Coll O, Patalano S, Gebauer F (2006) Drosophila UNR is required for translational repression of male-specific lethal 2 mRNA during regulation of X-chromosome dosage compensation. Genes & Development 20(3):380–389. doi:10.1101/gad.371906
Abbas YM, Pichlmair A, Gorna MW, Superti-Furga G, Nagar B (2013) Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature 494(7435):60–64. doi:10.1038/nature11783
Anantharaman V, Koonin EV, Aravind L (2002) Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Research 30(7):1427–1464
Antonicka H, Shoubridge EA (2015) Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Reports. doi:10.1016/j.celrep.2015.01.030
Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T (2012) Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdisciplinary Reviews RNA 3(2):159–177. doi:10.1002/wrna.1103
Auweter SD, Oberstrass FC, Allain FH (2006) Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Research 34(17):4943–4959. doi:10.1093/nar/gkl620
Balachandran S, Barber GN (2007) PKR in innate immunity, cancer, and viral oncolysis. Methods in Molecular Biology 383:277–301. doi:10.1007/978-1-59745-335-6_18
Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M et al (2012) The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Molecular Cell 46(5):674–690. doi:10.1016/j.molcel.2012.05.021
Barbalat R, Ewald SE, Mouchess ML, Barton GM (2011) Nucleic acid recognition by the innate immune system. Annual Review of Immunology 29:185–214. doi:10.1146/annurev-immunol-031210-101340
Barrandon C, Spiluttini B, Bensaude O (2008) Non-coding RNAs regulating the transcriptional machinery. Biology of the Cell/Under the Auspices of the European Cell Biology Organization 100(2):83–95. doi:10.1042/BC20070090
Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR (2005) 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 11(5):774–784. doi:10.1261/rna.7286705
Beckmann BM, Hoch PG, Marz M, Willkomm DK, Salas M, Hartmann RK (2012) A pRNA-induced structural rearrangement triggers 6S-1 RNA release from RNA polymerase in Bacillus subtilis. The EMBO Journal 31(7):1727–1738. doi:10.1038/emboj.2012.23
Beckmann BM, Horos R, Fischer B, Castello A, Eichelbaum K, Alleaume AM, Schwarzl T, Curk T, Foehr S, Huber W et al (2015) The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nature Communications 6:10127. doi:10.1038/ncomms10127
Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M (2011) The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334(6062):1524–1529. doi:10.1126/science.1212642
Brodersen DE, Clemons WM Jr, Carter AP, Wimberly BT, Ramakrishnan V (2002) Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. Journal of Molecular Biology 316(3):725–768. doi:10.1006/jmbi.2001.5359
Calabretta S, Richard S (2015) Emerging roles of disordered sequences in RNA-binding proteins. Trends in Biochemical Sciences 40(11):662–672. doi:10.1016/j.tibs.2015.08.012
Carlomagno T (2014) Present and future of NMR for RNA-protein complexes: a perspective of integrated structural biology. Journal of Magnetic Resonance 241:126–136. doi:10.1016/j.jmr.2013.10.007
Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM et al (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149(6):1393–1406. doi:10.1016/j.cell.2012.04.031
Castello A, Fischer B, Hentze MW, Preiss T (2013) RNA-binding proteins in Mendelian disease. Trends in Genetics: TIG 29(5):318–327. doi:10.1016/j.tig.2013.01.004
Castello A, Hentze MW, Preiss T (2015) Metabolic enzymes enjoying new partnerships as RNA-binding proteins. Trends in Endocrinology and Metabolism: TEM 26(12):746–757. doi:10.1016/j.tem.2015.09.012
Chang CH, Curtis JD, Maggi LB Jr, Faubert B, Villarino AV, O’Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J et al (2013) Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153(6):1239–1251. doi:10.1016/j.cell.2013.05.016
Charpentier E, Marraffini LA (2014) Harnessing CRISPR-Cas9 immunity for genetic engineering. Current Opinion in Microbiology 19:114–119. doi:10.1016/j.mib.2014.07.001
Choudhury NR, Nowak JS, Zuo J, Rappsilber J, Spoel SH, Michlewski G (2014) Trim25 is an RNA-specific activator of Lin28a/TuT4-mediated uridylation. Cell Reports 9(4):1265–1272. doi:10.1016/j.celrep.2014.10.017
Ciesla J (2006) Metabolic enzymes that bind RNA: yet another level of cellular regulatory network? Acta Biochimica Polonica 53(1):11–32
Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Molecular Cell 33(6):717–726. doi:10.1016/j.molcel.2009.01.026
Cooper TA, Wan L, Dreyfuss G (2009) RNA and disease. Cell 136(4):777–793. doi:10.1016/j.cell.2009.02.011
Dabo S, Meurs EF (2012) dsRNA-dependent protein kinase PKR and its role in stress, signaling and HCV infection. Viruses 4(11):2598–2635. doi:10.3390/v4112598
Darnell RB (2010) RNA regulation in neurologic disease and cancer. Cancer Research and Treatment 42(3):125–129. doi:10.4143/crt.2010.42.3.125
Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB (2001) Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107(4):489–499
Daubner GM, Clery A, Allain FH (2013) RRM-RNA recognition: NMR or crystallography…and new findings. Current Opinion in Structural Biology 23(1):100–108. doi:10.1016/j.sbi.2012.11.006
Donnelly N, Gorman AM, Gupta S, Samali A (2013) The eIF2alpha kinases: their structures and functions. Cellular and Molecular Life Sciences: CMLS 70(19):3493–3511. doi:10.1007/s00018-012-1252-6
Doudna JA, Sarnow P (2007) Translation initiation by viral internal ribosome entry sites. In: Mathews MB, Sonenberg N, Hershey JWB (eds) Translational control in biology and medicine, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 155–172
Duncan K, Grskovic M, Strein C, Beckmann K, Niggeweg R, Abaza I, Gebauer F, Wilm M, Hentze MW (2006) Sex-lethal imparts a sex-specific function to UNR by recruiting it to the msl-2 mRNA 3′ UTR: translational repression for dosage compensation. Genes & Development 20(3):368–379. doi:10.1101/gad.371406
ElAntak L, Tzakos AG, Locker N, Lukavsky PJ (2007) Structure of eIF3b RNA recognition motif and its interaction with eIF3j: structural insights into the recruitment of eIF3b to the 40 S ribosomal subunit. The Journal of Biological Chemistry 282(11):8165–8174. doi:10.1074/jbc.M610860200
Elroy-Stein O, Merrick WC (2007) Translation initiation via cellular internal ribosome entry sites. In: Mathews MB, Sonenberg N, Hershey JWB (eds) Translational control in biology and medicine, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 155–172
Forch P, Merendino L, Martinez C, Valcarcel J (2001) Modulation of msl-2 5′ splice site recognition by sex-lethal. RNA 7(9):1185–1191
Gawande B, Robida MD, Rahn A, Singh R (2006) Drosophila sex-lethal protein mediates polyadenylation switching in the female germline. The EMBO Journal 25(6):1263–1272. doi:10.1038/sj.emboj.7601022
Gebhardt A, Habjan M, Benda C, Meiler A, Haas DA, Hein MY, Mann A, Mann M, Habermann B, Pichlmair A (2015) mRNA export through an additional cap-binding complex consisting of NCBP1 and NCBP3. Nature Communications 6:8192. doi:10.1038/ncomms9192
Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW (2009) Disassembly of exon junction complexes by PYM. Cell 137(3):536–548. doi:10.1016/j.cell.2009.02.042
Gerstberger S, Hafner M, Tuschl T (2014) A census of human RNA-binding proteins. Nature Reviews Genetics 15(12):829–845. doi:10.1038/nrg3813
Ghezzi D, Saada A, D’Adamo P, Fernandez-Vizarra E, Gasparini P, Tiranti V, Elpeleg O, Zeviani M (2008) FASTKD2 nonsense mutation in an infantile mitochondrial encephalomyopathy associated with cytochrome C oxidase deficiency. American Journal of Human Genetics 83(3):415–423. doi:10.1016/j.ajhg.2008.08.009
Glisovic T, Bachorik JL, Yong J, Dreyfuss G (2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS letters 582(14):1977–1986. doi:10.1016/j.febslet.2008.03.004
Gold L, Janjic N, Jarvis T, Schneider D, Walker JJ, Wilcox SK, Zichi D (2012) Aptamers and the RNA world, past and present. Cold Spring Harbor Perspectives in Biology: 4(3): doi:10.1101/cshperspect.a003582
Graindorge A, Carre C, Gebauer F (2013) Sex-lethal promotes nuclear retention of msl2 mRNA via interactions with the STAR protein HOW. Genes & Development 27(12):1421–1433. doi:10.1101/gad.214999.113
Greenberg JR (1979) Ultraviolet light-induced crosslinking of mRNA to proteins. Nucleic Acids Research 6(2):715–732
Habchi J, Tompa P, Longhi S, Uversky VN (2014) Introducing protein intrinsic disorder. Chemical Reviews 114(13):6561–6588. doi:10.1021/cr400514h
Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G et al (2012) Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149(4):768–779. doi:10.1016/j.cell.2012.04.016
Hennig J, Militti C, Popowicz GM, Wang I, Sonntag M, Geerlof A, Gabel F, Gebauer F, Sattler M (2014) Structural basis for the assembly of the Sxl-Unr translation regulatory complex. Nature 515(7526):287–290. doi:10.1038/nature13693
Hennig J, Sattler M (2015) Deciphering the protein-RNA recognition code: combining large-scale quantitative methods with structural biology. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology 37(8):899–908. doi:10.1002/bies.201500033
Hentze MW, Preiss T (2010) The REM phase of gene regulation. Trends in Biochemical Sciences 35(8):423–426. doi:10.1016/j.tibs.2010.05.009
Heym RG, Niessing D (2012) Principles of mRNA transport in yeast. Cellular and Molecular Life Sciences: CMLS 69(11):1843–1853. doi:10.1007/s00018-011-0902-4
Hockensmith JW, Kubasek WL, Vorachek WR, von Hippel PH (1986) Laser cross-linking of nucleic acids to proteins. Methodology and first applications to the phage T4 DNA replication system. The Journal of Biological Chemistry 261(8):3512–3518
Hyman AA, Weber CA, Julicher F (2014) Liquid-liquid phase separation in biology. Annual Review of Cell and Developmental Biology 30:39–58. doi:10.1146/annurev-cellbio-100913-013325
Jia J, Arif A, Ray PS, Fox PL (2008) WHEP domains direct noncanonical function of glutamyl-prolyl tRNA synthetase in translational control of gene expression. Molecular Cell 29(6):679–690. doi:10.1016/j.molcel.2008.01.010
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821. doi:10.1126/science.1225829
Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J et al (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149(4):753–767. doi:10.1016/j.cell.2012.04.017
Kawai T, Akira S (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature Immunology 11(5):373–384. doi:10.1038/ni.1863
Keene JD (2007) RNA regulons: coordination of post-transcriptional events. Nature Reviews Genetics 8(7):533–543. doi:10.1038/nrg2111
Khawaja A, Vopalensky V, Pospisek M (2015) Understanding the potential of hepatitis C virus internal ribosome entry site domains to modulate translation initiation via their structure and function. Wiley Interdisciplinary Reviews RNA 6(2):211–224. doi:10.1002/wrna.1268
Kielkopf CL, Lucke S, Green MR (2004) U2AF homology motifs: protein recognition in the RRM world. Genes & Development 18(13):1513–1526. doi:10.1101/gad.1206204
Klein DJ, Moore PB, Steitz TA (2004) The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. Journal of Molecular Biology 340(1):141–177. doi:10.1016/j.jmb.2004.03.076
Kung JT, Colognori D, Lee JT (2013) Long noncoding RNAs: past, present, and future. Genetics 193(3):651–669. doi:10.1534/genetics.112.146704
Kwon SC, Yi H, Eichelbaum K, Fohr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN (2013) The RNA-binding protein repertoire of embryonic stem cells. Nature Structural & Molecular Biology 20(9):1122–1130. doi:10.1038/nsmb.2638
Le Hir H, Sauliere J, Wang Z (2016) The exon junction complex as a node of post-transcriptional networks. Nature Reviews Molecular Cell Biology 17(1):41–54. doi:10.1038/nrm.2015.7
Lee I, Hong W (2004) RAP—a putative RNA-binding domain. Trends in Biochemical Sciences 29(11):567–570. doi:10.1016/j.tibs.2004.09.005
Li S, Wang L, Fu B, Dorf ME (2014) Trim65: a cofactor for regulation of the microRNA pathway. RNA Biology 11(9):1113–1121. doi:10.4161/rna.36179
Lin Y, Protter DS, Rosen MK, Parker R (2015) Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Molecular Cell 60(2):208–219. doi:10.1016/j.molcel.2015.08.018
Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB (2003) Protein disorder prediction: implications for structural proteomics. Structure 11(11):1453–1459
Loedige I, Gaidatzis D, Sack R, Meister G, Filipowicz W (2013) The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Research 41(1):518–532. doi:10.1093/nar/gks1032
Loedige I, Jakob L, Treiber T, Ray D, Stotz M, Treiber N, Hennig J, Cook KB, Morris Q, Hughes TR et al (2015) The crystal structure of the NHL domain in complex with RNA reveals the molecular basis of Drosophila brain-tumor-mediated gene regulation. Cell Reports 13(6):1206–1220. doi:10.1016/j.celrep.2015.09.068
Loedige I, Stotz M, Qamar S, Kramer K, Hennig J, Schubert T, Loffler P, Langst G, Merkl R, Urlaub H et al (2014) The NHL domain of BRAT is an RNA-binding domain that directly contacts the hunchback mRNA for regulation. Genes & Development 28(7):749–764. doi:10.1101/gad.236513.113
Lomakin IB, Hellen CU, Pestova TV (2000) Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Molecular and Cellular Biology 20(16):6019–6029
Luirink J, Sinning I (2004) SRP-mediated protein targeting: structure and function revisited. Biochimica et Biophysica Acta 1694(1-3):17–35. doi:10.1016/j.bbamcr.2004.03.013
Lukavsky PJ (2009) Structure and function of HCV IRES domains. Virus Research 139(2):166–171. doi:10.1016/j.virusres.2008.06.004
Lukong KE, Chang KW, Khandjian EW, Richard S (2008) RNA-binding proteins in human genetic disease. Trends in Genetics: TIG 24(8):416–425. doi:10.1016/j.tig.2008.05.004
Lunde BM, Moore C, Varani G (2007) RNA-binding proteins: modular design for efficient function. Nature Reviews Molecular Cell Biology 8(6):479–490. doi:10.1038/nrm2178
Marraffini LA (2015) CRISPR-Cas immunity in prokaryotes. Nature 526(7571):55–61. doi:10.1038/nature15386
Matia-Gonzalez AM, Laing EE, Gerber AP (2015) Conserved mRNA-binding proteomes in eukaryotic organisms. Nature Structural & Molecular Biology 22(12):1027–1033. doi:10.1038/nsmb.3128
Meister G (2013) Argonaute proteins: functional insights and emerging roles. Nature Reviews Genetics 14(7):447–459. doi:10.1038/nrg3462
Mili S, Steitz JA (2004) Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 10(11):1692–1694. doi:10.1261/rna.7151404
Mitchell SF, Jain S, She M, Parker R (2013) Global analysis of yeast mRNPs. Nature Structural & Molecular Biology 20(1):127–133. doi:10.1038/nsmb.2468
Mitchell SF, Parker R (2014) Principles and properties of eukaryotic mRNPs. Molecular Cell 54(4):547–558. doi:10.1016/j.molcel.2014.04.033
Mondragon A (2013) Structural studies of RNase P. Annual Review of Biophysics 42:537–557. doi:10.1146/annurev-biophys-083012-130406
Morris AR, Mukherjee N, Keene JD (2010) Systematic analysis of posttranscriptional gene expression. Wiley Interdisciplinary Reviews Systems Biology and Medicine 2(2):162–180. doi:10.1002/wsbm.54
Muckenthaler MU, Galy B, Hentze MW (2008) Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annual Review of Nutrition 28:197–213. doi:10.1146/annurev.nutr.28.061807.155521
Mukhopadhyay R, Jia J, Arif A, Ray PS, Fox PL (2009) The GAIT system: a gatekeeper of inflammatory gene expression. Trends in Biochemical Sciences 34(7):324–331. doi:10.1016/j.tibs.2009.03.004
Muller M, Heym RG, Mayer A, Kramer K, Schmid M, Cramer P, Urlaub H, Jansen RP, Niessing D (2011) A cytoplasmic complex mediates specific mRNA recognition and localization in yeast. PLoS Biology 9(4):e1000611. doi:10.1371/journal.pbio.1000611
Neelamraju Y, Hashemikhabir S, Janga SC (2015) The human RBPome: from genes and proteins to human disease. Journal of Proteomics 127(Pt A):61–70. doi:10.1016/j.jprot.2015.04.031
Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD et al (2015) Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Molecular Cell 57(5):936–947. doi:10.1016/j.molcel.2015.01.013
Nudler E, Mironov AS (2004) The riboswitch control of bacterial metabolism. Trends in Biochemical Sciences 29(1):11–17. doi:10.1016/j.tibs.2003.11.004
Oldenburg M, Kruger A, Ferstl R, Kaufmann A, Nees G, Sigmund A, Bathke B, Lauterbach H, Suter M, Dreher S et al (2012) TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science 337(6098):1111–1115. doi:10.1126/science.1220363
Pashev IG, Dimitrov SI, Angelov D (1991) Crosslinking proteins to nucleic acids by ultraviolet laser irradiation. Trends in Biochemical Sciences 16(9):323–326
Perard J, Rasia R, Medenbach J, Ayala I, Boisbouvier J, Drouet E, Baudin F (2009) Human initiation factor eIF3 subunit b interacts with HCV IRES RNA through its N-terminal RNA recognition motif. FEBS letters 583(1):70–74. doi:10.1016/j.febslet.2008.11.025
Pestova TV, Hellen CU, Shatsky IN (1996) Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Molecular and Cellular Biology 16(12):6859–6869
Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D et al (2011) Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nature Structural & Molecular Biology 18(7):796–804. doi:10.1038/nsmb.2064
Popow J, Alleaume AM, Curk T, Schwarzl T, Sauer S, Hentze MW (2015) FASTKD2 is an RNA-binding protein required for mitochondrial RNA processing and translation. RNA 21(11):1873–1884. doi:10.1261/rna.052365.115
Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, Fox PL (2009) A stress-responsive RNA switch regulates VEGFA expression. Nature 457(7231):915–919. doi:10.1038/nature07598
Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A et al (2013) A compendium of RNA-binding motifs for decoding gene regulation. Nature 499(7457):172–177. doi:10.1038/nature12311
Ricci EP, Kucukural A, Cenik C, Mercier BC, Singh G, Heyer EE, Ashar-Patel A, Peng L, Moore MJ (2014) Staufen1 senses overall transcript secondary structure to regulate translation. Nature Structural & Molecular Biology 21(1):26–35. doi:10.1038/nsmb.2739
Riley KJ, Steitz JA (2013) The “observer effect” in genome-wide surveys of protein-RNA interactions. Molecular Cell 49(4):601–604. doi:10.1016/j.molcel.2013.01.030
Riley KJ, Yario TA, Steitz JA (2012) Association of argonaute proteins and microRNAs can occur after cell lysis. RNA 18(9):1581–1585. doi:10.1261/rna.034934.112
Safaee N, Kozlov G, Noronha AM, Xie J, Wilds CJ, Gehring K (2012) Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Molecular Cell 48(3):375–386. doi:10.1016/j.molcel.2012.09.001
Scherrer T, Mittal N, Janga SC, Gerber AP (2010) A screen for RNA-binding proteins in yeast indicates dual functions for many enzymes. PLoS One 5(11):e15499. doi:10.1371/journal.pone.0015499
Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E (2005) Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310(5753):1513–1515. doi:10.1126/science.1118977
Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU (1998) Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. Journal of Virology 72(6):4775–4782
Sternberg SH, Doudna JA (2015) Expanding the biologist’s toolkit with CRISPR-Cas9. Molecular Cell 58(4):568–574. doi:10.1016/j.molcel.2015.02.032
Strein C, Alleaume AM, Rothbauer U, Hentze MW, Castello A (2014) A versatile assay for RNA-binding proteins in living cells. RNA 20(5):721–731. doi:10.1261/rna.043562.113
Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D’Ambrogio A, Luscombe NM, Ule J (2015) hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 519(7544):491–494. doi:10.1038/nature14280
Sun C, Querol-Audi J, Mortimer SA, Arias-Palomo E, Doudna JA, Nogales E, Cate JH (2013) Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Nucleic Acids Research 41(15):7512–7521. doi:10.1093/nar/gkt510
Thandapani P, O’Connor TR, Bailey TL, Richard S (2013) Defining the RGG/RG motif. Molecular Cell 50(5):613–623. doi:10.1016/j.molcel.2013.05.021
Tsvetanova NG, Klass DM, Salzman J, Brown PO (2010) Proteome-wide search reveals unexpected RNA-binding proteins in Saccharomyces cerevisiae. PLoS One: 5(9): doi:10.1371/journal.pone.0012671
Vuzman D, Levy Y (2010) DNA search efficiency is modulated by charge composition and distribution in the intrinsically disordered tail. Proceedings of the National Academy of Sciences of the United States of America 107(49):21004–21009. doi:10.1073/pnas.1011775107
Vuzman D, Levy Y (2012) Intrinsically disordered regions as affinity tuners in protein-DNA interactions. Molecular Biosystems 8(1):47–57. doi:10.1039/c1mb05273j
Walden WE, Selezneva AI, Dupuy J, Volbeda A, Fontecilla-Camps JC, Theil EC, Volz K (2006) Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science 314(5807):1903–1908. doi:10.1126/science.1133116
Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Molecular Cell 43(6):904–914. doi:10.1016/j.molcel.2011.08.018
Wang X, McLachlan J, Zamore PD, Hall TM (2002) Modular recognition of RNA by a human pumilio-homology domain. Cell 110(4):501–512
Wang Y, Wang Z, Tanaka Hall TM (2013) Engineered proteins with Pumilio/fem-3 mRNA binding factor scaffold to manipulate RNA metabolism. The FEBS Journal 280(16):3755–3767. doi:10.1111/febs.12367
Wassarman KM, Storz G (2000) 6S RNA regulates E. coli RNA polymerase activity. Cell 101(6):613–623
Watkins NJ, Bohnsack MT (2012) The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdisciplinary Reviews RNA 3(3):397–414. doi:10.1002/wrna.117
Weber SC, Brangwynne CP (2012) Getting RNA and protein in phase. Cell 149(6):1188–1191. doi:10.1016/j.cell.2012.05.022
Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E (2016) Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science: 351(6270): doi:10.1126/science.aad4939
Will CL, Luhrmann R (2011) Spliceosome structure and function. Cold Spring Harbor Perspectives in Biology: 3(7): doi:10.1101/cshperspect.a003707
Yamamoto H, Collier M, Loerke J, Ismer J, Schmidt A, Hilal T, Sprink T, Yamamoto K, Mielke T, Burger J et al (2015) Molecular architecture of the ribosome-bound hepatitis C virus internal ribosomal entry site RNA. The EMBO Journal 34(24):3042–3058. doi:10.15252/embj.201592469
Yao P, Potdar AA, Ray PS, Eswarappa SM, Flagg AC, Willard B, Fox PL (2013) The HILDA complex coordinates a conditional switch in the 3′-untranslated region of the VEGFA mRNA. PLoS Biology 11(8):e1001635. doi:10.1371/journal.pbio.1001635
Yu M, Levine SJ (2011) Toll-like receptor, RIG-I-like receptors and the NLRP3 inflammasome: key modulators of innate immune responses to double-stranded RNA viruses. Cytokine & Growth Factor Reviews 22(2):63–72. doi:10.1016/j.cytogfr.2011.02.001
Zappulla DC, Cech TR (2006) RNA as a flexible scaffold for proteins: yeast telomerase and beyond. Cold Spring Harbor Symposia on Quantitative Biology 71:217–224. doi:10.1101/sqb.2006.71.011
Zhang Q, Kim NK, Feigon J (2011) Architecture of human telomerase RNA. Proceedings of the National Academy of Sciences of the United States of America 108(51):20325–20332. doi:10.1073/pnas.1100279108
Acknowledgments
BB acknowledges the funding from the German Research Foundation (DFG grant ZUK75/1). AC is funded by the MRC Career Development Award no. MR/L019434/1. JM acknowledges the funding from the Bavarian Research Network for Molecular Biosystems (BioSysNet), the German Federal Ministry of Education and Research (BMBF, SUPR-G, grant 01ZX1401D) and the German Research Foundation (DFG, grant ME4238/1-1). We are grateful to Leonhard Jakob for the help in preparation of Fig. 2. We would also like to acknowledge the two anonymous reviewers for their constructive criticism and valuable comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Beckmann, B.M., Castello, A. & Medenbach, J. The expanding universe of ribonucleoproteins: of novel RNA-binding proteins and unconventional interactions. Pflugers Arch - Eur J Physiol 468, 1029–1040 (2016). https://doi.org/10.1007/s00424-016-1819-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00424-016-1819-4