Abstract
Purpose
Circulating testosterone levels are a heritable trait with anabolic properties in various tissues, including skeletal muscle. So far, hundreds of single nucleotide polymorphisms (SNPs) associated with testosterone levels have been identified in nonathletic populations. The aim of the present study was to test the association of 822 testosterone-increasing SNPs with muscle-related traits (muscle fiber size, fat-free mass and handgrip strength) and to validate the identified SNPs in independent cohorts of strength and power athletes.
Methods
One hundred and forty-eight physically active individuals (47 females, 101 males) were assessed for cross-sectional area (CSA) of fast-twitch muscle fibers. Significant SNPs were further assessed for fat-free mass and handgrip strength in > 354,000 participants from the UK Biobank cohort. The validation cohorts included Russian elite athletes.
Results
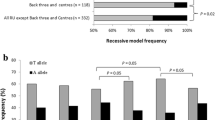
From an initial panel of 822 SNPs, we identified five testosterone-increasing alleles (DOCK3 rs77031559 G, ESR1 rs190930099 G, GLIS3 rs34706136 TG, GRAMD1B rs850294 T, TRAIP rs62260729 C) nominally associated (P < 0.05) with CSA of fast-twitch muscle fibers, fat-free mass and handgrip strength. Based on these five SNPs, the number of testosterone-increasing alleles was positively associated with testosterone levels in male athletes (P = 0.048) and greater strength performance in weightlifters (P = 0.017). Moreover, the proportion of participants with ≥ 2 testosterone-increasing alleles was higher in power athletes compared to controls (68.9 vs. 55.6%; P = 0.012).
Conclusion
Testosterone-related SNPs are associated with muscle fiber size, fat-free mass and strength, which combined can partially contribute to a greater predisposition to strength/power sports.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Testosterone is an anabolic–androgenic steroid hormone produced mainly in Leydig cells of the testes in men and the ovary and the adrenal cortex in women. Testosterone plays an integral role in the development and maintenance of male characteristics, including the development of primary and secondary sex characteristics and the maintenance of the reproductive system. In addition, testosterone plays a clear role on several non-reproductive tissues, regardless of gender. In skeletal muscle, testosterone and its metabolite, dihydrotestosterone, have a well-defined anabolic property, mainly through an increase in protein synthesis via the activation of the mammalian target of rapamycin (mTOR) pathway together with the androgen receptor (AR) signaling (Basualto-Alarcon et al. 2013; Zeng et al. 2017). Other anabolic or anti-catabolic mechanisms have also been proposed (Dubois et al. 2012), all suggestive that testosterone plays an important role in muscle mass regulation.
Testosterone administration has been shown to increase muscle mass and strength in a dose-dependent manner in young and older men (Bhasin et al. 2001; Bhasin et al. 2005) and in young women (Horwath et al. 2020). Given that muscle hypertrophy (and the increase in muscle function it brings) has a performance-enhancing effect in sports that depend on strength and power, higher levels of testosterone create an advantage (Wood and Stanton 2012). Not surprisingly, testosterone is the most common form of doping in sport; however, it should be mentioned that due to the dynamic regulation of its endogenous production, testosterone concentrations may vary considerably within and among individuals. There is a strong heritability for serum testosterone, with genetic factors accounting for 40–70% of the variation in testosterone levels in men (Travison et al. 2014) and 65% in women (Hong et al. 2001).
It is possible that individuals who have higher levels of endogenous testosterone are more predisposed to certain power sports. In an assessment of a large cohort of elite male athletes, sprinters showed higher free testosterone levels than athletes in other sports (e.g., long-distance runners) (Bermon and Garnier 2017). Similarly, in an assessment of a large cohort of elite female athletes, sprinters showed higher testosterone levels than long-distance runners (Bermon et al. 2014). Higher testosterone levels in female sprinters can contribute to athletic success, allowing them to reach a higher competitive level (Ahmetov et al. 2020). Indeed, female athletes with higher free testosterone performed better in 400 to 800-m sprinting events, hammer throw and pole vault compared with female competitors with lower free testosterone (Bermon and Garnier 2017). Testosterone is a contributory trait in the complex nature of athletic phenotypes, and can influence athletic performance (e.g., increased neuronal activity, bone growth and hemoglobin levels) (Wood and Stanton 2012).
A recent genome-wide association study (GWAS) provided a number of single nucleotide polymorphisms (SNPs) associated with higher total and bioavailable testosterone levels in men and women (Ruth et al. 2020). These innate characteristics may lead to interindividual differences in hormone levels capable of influencing testosterone-related phenotypes, such as predisposition to increase muscle mass and strength. However, with the exception of a polymorphism in the AR gene (with the potential to affect testosterone levels) (Guilherme et al. 2021b), the interaction of testosterone-increasing alleles with muscle mass and function remains to be investigated.
Testosterone-induced gains in muscle size were associated with a significant increase in muscle fiber cross-sectional area (CSA). In young healthy, eugonadal men treated with graded doses of testosterone, the increases in muscle volume are associated with concentration-dependent increases in CSA of both type I and type II muscle fibers (Sinha-Hikim et al. 2002). In young healthy, physically active women (20–35 years) treated with testosterone cream for 10 weeks, muscle hypertrophy was primarily driven by increases in CSA of type II fibers (Horwath et al. 2020). Overall, the CSA of muscle fibers correlates positively with strength variables, especially when it comes to type II (fast-twitch) fibers. Since fast-twitch fibers are required in high-energy movement tasks such as sprinting or weightlifting, their CSA is of vital importance for power athletes.
Muscle CSA can be affected by numerous environmental factors, but it is also highly determined by genetic factors. One can inherit genetic polymorphisms which make muscle hypertrophy easier than others who do not possess those polymorphisms. Some individual SNPs were associated with larger CSA of fast-twitch muscle fibers (Ahmetov et al. 2008; Broos et al. 2016; Grishina et al. 2019; Seaborne et al. 2019). These SNPs, associated with a larger CSA of fast-twitch fibers, were also more frequent in strength and power athletes, which suggest a favourable genetic profile. Athletic phenotypes (including muscle strength and power) are polygenic in nature, which implies that multiple polymorphisms influence these athletic phenotype (Guilherme and Lancha 2020; Moreland et al. 2020).
Although there is a relationship between testosterone levels and muscle mass regulation, the shared genetic background between testosterone-increasing alleles and muscle CSA is poorly understood. Therefore, the purpose of the present study was to explore whether GWAS-identified testosterone-increasing SNPs (Ruth et al. 2020) were associated with muscle fiber size (CSA of fast-twitch muscle fibers), fat-free mass and muscle strength. To validate the SNPs that met the selection criteria (testosterone-increasing alleles associated with muscle-related traits), independent athlete cohorts were assessed using a polygenic approach. The combined association of the selected SNPs (based on the number of favorable alleles) was assessed for testosterone levels in male athletes, strength performance in elite weightlifters and prevalence in strength and power athletes.
Materials and methods
Participants and ethical approval
The analysis of the CSA of fast-twitch muscle fibers was carried out in 148 physically active participants with mixed training (i.e., aerobic + resistance) background (Table 1). They were classified according to their training frequency as mildly active (2 training sessions per week), moderately active (3–4 training sessions per week), highly active (5–7 training sessions per week) or extremely active (two training sessions per day). Testosterone-increasing SNPs associated with the CSA of fast-twitch muscle fibers were subsequently tested for associations with fat-free mass and handgrip strength in the UK Biobank—a prospective population-based study of > 354,000 individuals (summary statistics is available from https://genetics.opentargets.org/).
Following this two-stage association analysis, the selected SNPs (using a polygenic approach) were tested in three independent athlete cohorts: (i) 49 male athletes with power component in their athletic performance (age 22.8 ± 0.7 years; 1 100-m runner, 4 badminton players, 7 baseball players, 4 boxers, 5 canoeists, 8 figure skaters, 2 kayakers, 6 rowers, 3 speed skaters, 4 volleyball players and 5 wrestlers) were tested for testosterone levels; (ii) 50 weightlifters (29 males, age 23.7 ± 0.7 years; 21 females, age 22.5 ± 0.8 years) were tested for strength performance in official competitions; (iii) 222 international-level power athletes (126 males, age 23.6 ± 0.4 years; 96 females, age 22.8 ± 0.5 years) were tested for the prevalence of favorable alleles compared to 151 non-athlete controls (120 males and 31 females, age 44.7 ± 4.0 years). The power athlete group was composed of 67 strength athletes (17 powerlifters and 50 weightlifters), 82 speed-strength athletes (15 alpine skiers, 3 arm-wrestlers, 29 climbers, 4 heptathletes and decathletes, 22 jumpers and 9 throwers) and 73 sprinters (25 100- to 400-m runners, 19 200- to 500-m kayakers and 29 500- to 1000-m speed skaters). Athletes were Russian national team members who had never tested positive for doping. Controls were healthy unrelated Russians without any competitive sport experience.
A flow diagram displaying the selection process of the significant SNPs is shown in Fig. 1. This study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of the Federal Research and Clinical Center of Physical–chemical Medicine. Written informed consent was obtained from each participant.
Muscle biopsy and determination of the CSA of fast-twitch muscle fibers
Vastus lateralis samples of 148 physically active participants were obtained from the left leg using the modified Bergström needle procedure with aspiration under local anaesthesia with 2% lidocaine solution. Prior to analysis, samples were frozen in liquid nitrogen and stored at − 80 °C. Serial cross-sections (7 μm) were obtained from frozen samples using an ultratom (Leica Microsystems, Germany). Sections were thaw-mounted on Polysine glass slides, maintained at room temperature (RT) for 15 min and incubated in PBS (3 × 5 min). The sections were then incubated at RT in primary antibodies against slow or fast isoforms of the myosin heavy chains (M8421, 1:5000; M4276; 1:600, respectively; Sigma-Aldrich, USA) for 1 h and incubated in PBS (3 × 5 min). Next, the sections were incubated at RT in secondary antibodies conjugated with FITC (F0257; 1:100; Sigma-Aldrich) for 1 h. The antibodies were removed, and the sections washed in PBS (3 × 5 min), placed in mounting media and covered with a cover slip. Images were captured by fluorescent microscope (Eclipse Ti-U, Nikon, Japan). All analyzed images contained 329 ± 14 fibers. The ratio of the number of stained fibers to the total fiber number was calculated. Fibers stained in serial sections with antibodies against slow and fast isoforms were considered hybrid fibers.
Hormone levels and strength performance assessment
Resting testosterone levels were examined in serum of athletes. A total of 10 mL of venous blood were collected the morning after an overnight fast and sleep in tubes containing EDTA and placed at 4 °C until processing (blood was collected at least 15 h after the last training). Testosterone was analyzed on a microplate spectrophotometer (Bio-Rad, Hercules, CA, USA) using an enzyme immunoassay test (Alkor-Bio, St Petersburg, Russia).
Evaluation of strength in weightlifters was computed based on their performance in the snatch, and clean and jerk (best results in official competitions, including Olympic Games, European and World Championships). The total weight lifted (in kg) is multiplied by the Wilks Coefficient (Coeff) to find the standard amount lifted normalized across all body weights, as previously described (Grishina et al. 2019).
DNA extraction and genotyping
Molecular genetic analysis was performed with DNA samples obtained from leukocytes (venous blood). DNA extraction and purification were performed using a commercial kit according to the manufacturer’s instructions (Technoclon, Moscow, Russia). Genotyping of SNPs was majorly performed using microarray technology, as previously described (Guilherme et al. 2021a).
Statistical analyses
Statistical analyses were conducted using PLINK 1.9, R (3.4.3), and GraphPad InStat (GraphPad Software, Inc., USA). Haplotype phasing before imputation was performed using SHAPEIT. Imputation was performed using IMPUTE2. For phasing and imputation, we used 1000 Genomes Phase 3 data as a reference panel and imputed the variants with a frequency higher than 0.1% in the reference panel. Variants imputed with low certainty (info score < 0.6) were filtered out after imputation. Pearson’s correlation assessed the relationships between the number of favorable alleles (polygenic analysis) and different phenotypes. Allele frequencies between athletes and controls were compared using χ2 tests. All calculations were performed adjusting for covariates (muscle fiber size: principal component analysis (PCA), age, sex, physical activity and training type; handgrip strength: PCA, age, sex; fat-free mass: PCA, age, sex; testosterone levels in men: age; strength performance in weightlifters: age, sex). In particular, the search for association between 822 SNPs and CSA of fast-twitch muscle fibers was performed using logistic regression analysis adjusted for covariates. Data of testosterone levels were used from the study involving 425,097 UK Biobank participants by Ruth et al. (2020). Data of handgrip strength were used from the study involving 359,729 UK Biobank participants. Data of fat-free mass were used from the study involving 354,808 UK Biobank participants. Summary statistics for handgrip strength and fat-free mass are available from https://genetics.opentargets.org/. P values < 0.05 were considered statistically significant.
Results
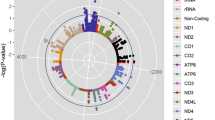
In the first stage, we tested the hypothesis that genome-wide significant testosterone-increasing alleles are associated with muscle fiber size. For this, we selected a panel of 855 SNPs (Supplementary Table 1) from the study by Ruth et al. (Ruth et al. 2020). However, the set of testosterone-increasing alleles included 822 SNPs, because 33 SNPs had directionally opposing effects between sexes and were not included in the present study. Of the 822 SNPs, 31 were nominally associated (P < 0.05) with CSA of fast-twitch muscle fibers (adjusted for covariates), with the same direction of association (i.e., testosterone-increasing alleles were associated with increased CSA). After exclusion of SNPs localized in the X chromosome (only autosomal chromosomes were evaluated), the set of testosterone-increasing alleles associated with muscle fiber size included 28 SNPs (Supplementary Table 2).
Next, using UK Biobank database, we have identified that of the 28 SNPs, 5 testosterone-increasing alleles (DOCK3 rs77031559 G, ESR1 rs190930099 G, GLIS3 rs34706136 TG, GRAMD1B rs850294 T, TRAIP rs62260729 C) were also nominally associated (P < 0.05) with increased handgrip strength and fat-free mass in 359,729 and 354,808 individuals, respectively (Table 2).
Although only two associations (rs34706136 for handgrip strength and rs190930099 for fat-free mass) passed Bonferroni correction for multiple testing (i.e. P value = 0.05/822 SNPs * 3 traits (CSA of muscle fibers, handgrip strength, fat-free mass) = 0.00002), we felt justified to use all 5 SNPs in the polygenic analysis given that we used SNPs already discovered independently via GWAS of testosterone levels at genome-wide significance (Ruth et al. 2020). To validate the five SNPs associated with testosterone levels and muscle-related traits, we performed a series of studies in independent cohorts of athletes using a polygenic approach (i.e., the combined association of the 5 SNPs), as shown in Fig. 1. We classified all participants according to the number of testosterone-increasing alleles they possessed, that is, carriers of DOCK3 rs77031559 AA, ESR1 rs190930099 AA, GLIS3 rs34706136 TT, GRAMD1B rs850294 CC, and TRAIP rs62260729 TT genotypes had zero testosterone-increasing alleles, whereas participants with DOCK3 rs77031559 GG, ESR1 rs190930099 GG, GLIS3 rs34706136 TG/TG, GRAMD1B rs850294 TT, and TRAIP rs62260729 CC genotypes had 10 testosterone-increasing alleles (heterozygous genotypes were computed with intermediate scores).
The number of testosterone-increasing alleles was positively associated with testosterone levels in elite male athletes (r = 0.28; P = 0.048) and with greater strength performance (total lifts in snatch and clean and jerk adjusted for sex and weight) in elite weightlifters (r = 0.34; P = 0.017). None of the participants (athletes or controls) had the maximum number of testosterone-increasing alleles (range from 0 to 6 alleles). However, the proportion of participants with a high number of testosterone-increasing alleles (i.e., ≥ 2 alleles instead of 0–1 allele) was greater among power athletes compared to controls (68.9 vs 55.6%; odds ratio (OR) = 1.8, P = 0.012). Table 3 summarizes the findings of the polygenic analysis.
Discussion
To our knowledge, this is the first study aimed towards identifying the shared genetic background between testosterone-increasing alleles, muscle traits and athletic performance. We identified five SNPs that were associated with testosterone levels, CSA of fast-twitch muscle fibers, fat-free-mas and handgrip strength.
The five identified SNPs are located in introns of genes that have multiple functions in relation to the endocrine system, metabolism and cellular function. More specifically, DOCK3 (Dedicator Of Cytokinesis 3) encodes a protein involved in the regulation of actin cytoskeleton and cell adhesion receptors; ESR1 (Estrogen Receptor 1) encodes a nuclear receptor for estrogen that controls many cellular processes including growth and differentiation; GLIS3 (GLIS Family Zinc Finger 3) encodes a protein involved in the development of pancreatic beta cells, the thyroid, liver and kidney; GRAMD1B (GRAM Domain Containing 1B) encodes a protein that plays a crucial role in cholesterol homeostasis; TRAIP (TRAF Interacting Protein) encodes a protein involved in cell activation and protection against apoptosis. Interestingly, in an assessment of young, healthy men who underwent 10 weeks of resistance training, three of these genes alter their expression in skeletal muscle in response to resistance training compared to pre-training (ESR1 and GLIS) or endurance training (GRAMD1B) (Vissing and Schjerling 2014). Moreover, according to the GTEx portal (https://gtexportal.org), two SNPs (TRAIP rs62260729 and DOCK3 rs77031559) are functional and influence the expression of several genes in various tissues, including testis, adrenal gland and skeletal muscle—all important in terms of testosterone production, physical performance and training responses. Noteworthy, TRAIP rs62260729 C allele is associated with increased expression of CDHR4 gene, which has its expression increased in response to resistance exercise (Vissing and Schjerling 2014). More details of gene function, effects of SNPs and gene expression following resistance training are shown in Supplementary Table 3.
The physiological implication of higher testosterone levels in skeletal muscle is the maintenance or increase (hypertrophy) of skeletal muscle mass, and a subsequent indirect increase in muscle strength, which can be advantageous for power athletes. Larger fast-twitch fibers lead to a larger whole muscle and a greater muscle volume. Top-level sprinters, for example, have a higher fat-free mass due to greater muscle volume, which can explain almost half (47.5%) of the variability in sprint performance (Miller et al. 2020). A larger muscle volume is able to generate stronger and more powerful contractions, resulting in greater sprint speed (Miller et al. 2020). Although it is well established that muscle volume can be affected by numerous environmental factors, genetic variability between individuals likely determines the extension of muscle adaptation. In the present study, those with ≥ 2 testosterone-increasing alleles (also associated with muscle fiber size, fat-free mass and handgrip strength) had greater strength (weightlifting) performance in competition and are 1.8 times more likely to be an elite power athlete. These 5 SNPs can be part of a favorable polygenic profile for muscle hypertrophy and strength performance (the innate predisposition to complex phenotypes involves the sum of several common polymorphisms). However, the biological role of these genes and SNPs in skeletal muscle is not fully understood.
The ESR1 gene is probably the one that has the most evidence in relation to skeletal muscle growth. Animal studies have shown that ESR1 elimination resulted in an increase in tibialis anterior muscle mass (a fast-twitch muscle) (Brown et al. 2009). In turn, an assessment of the pre-training skeletal muscle transcriptome of healthy men and women clustered as non-responders (Non), modest responders (Mod), and extreme responders (Xtr) to resistance training (based on differential magnitudes of myofiber hypertrophy), there was a stepwise increase in ESR1 expression from Non to Mod to Xtr, suggesting that estrogen signaling may be important for increased hypertrophic capacity (Thalacker-Mercer et al. 2013). Our findings support the relevance of the ESR1 gene for muscle hypertrophy, with the rs190930099 G allele likely playing a role. We did not assess the participants' estrogen levels, but a recent study showed that a cluster of testosterone-increasing alleles also increased estradiol levels in men (consistent with the physiological conversion of testosterone to estrogen) (Ruth et al. 2020). Of note, the ESR1 gene is expressed in skeletal muscle of men and women (Lemoine et al. 2003) and is therefore a tissue target for estrogen action.
Two other identified genes (CDHR4 [near TRAIP] and GLIS3) are also responsive to resistance training (Vissing and Schjerling, 2014). The CDHR4 gene [near TRAIP] was found to be necessary for axon guidance and cell migration in GABAergic neuromuscular junction development (Ackley 2014), which seems to play a signaling role in the contractile activity of skeletal muscle (Lenina et al. 2019). The GLIS3 gene have been associated with a decreased risk of knee osteoarthritis (Zhang et al. 2021), but its relationship with muscle performance remains to be further investigated. Although there is no evidence of gene regulation by resistance training, the DOCK3 may also be a contributing factor. The DOCK3 gene promotes axonal outgrowth via cytoskeleton reorganization and plays an important role in the muscle tone (Helbig et al. 2017). Furthermore, the DOCK family of proteins has been shown to bind to regulators of PTEN/AKT signaling (Jungmichel et al. 2014), an important signaling for muscle hypertrophy. Molecular inhibition of DOCK3 in skeletal muscle increases phosphorylated AKT levels, which influences the muscle morphology and function (Alexander et al. 2014). In general, the SNPs in the aforementioned genes were associated with muscle hypertrophy and strength, but still need to elucidate whether they are acting directly on skeletal muscle.
The only identified gene that has a known direct relationship to testosterone production was GRAMD1B, which belongs to a family of sterol-binding proteins. Steroidogenic cells take up cholesterol to initiate steroidogenesis (i.e., cholesterol is a substrate for testosterone biosynthesis), and the GRAMD1B gene assists in the transfer of cholesterol from the plasma membrane to the endoplasmic reticulum, where steroid hormones are produced (Larsen et al. 2020). GRAMD1 proteins facilitate the movement of accessible plasma membrane cholesterol to the endoplasmic reticulum, and cells that lack GRAMD1 proteins result in less efficient cholesterol transfer (Naito et al. 2019). It is unknown how the GRAMD1B rs850294 affects testosterone levels; however, this gene has been shown to be expressed differently between resistance and endurance training—opposite ends of the training-induced muscle adaptation continuum (Vissing and Schjerling 2014). It is well established that more intense exercises (common in a resistance training program) induce a greater increase in circulating testosterone levels (D'Andrea et al. 2020), as well as greater muscle hypertrophy (Lasevicius et al. 2018). In line with this, muscle power-related SNPs, such as ACTN3 R577X, have previously been associated with higher testosterone levels (Ahmetov et al. 2014; Pimenta et al. 2012).
It has been shown that there is a positive change in lean mass per unit higher of testosterone (bioavailable testosterone in men and testosterone total in women) (Ruth et al. 2020). Testosterone, like all other hormones, act in an integrated communication network responsible for modulating cellular signaling. Therefore, the combination of the 5 SNPs identified in this study could somehow favor the hypertrophy of fast-twitch muscle fibers and strength performance. Here, we explore the influence of these SNPs (under a polygenic profile) in the context of sporting excellence, however, the issue is also relevant for clinical conditions affecting muscle mass.
Our study does have limitations. First, none of the associations between SNPs and CSA of muscle fibers passed correction for multiple testing, but we felt justified to use five SNPs in the polygenic analysis given that we used SNPs which were initially found in GWAS, meaning that in the discovery phase (Ruth et al. 2020) these SNPs have passed correction for multiple testing at genome-wide significance (P < 5.0·10–8). It is common not to adjust for multiple comparisons in the validation phase to prevent the loss of potentially important findings (Duncan et al. 2019; Wood et al. 2014). Second, there may be other SNPs acting on the traits of interest that the present study was unable to detect. It is worth mentioning that this study was an initial approach to align a large number of testosterone-related SNPs with physiological and functional data in elite athletes. Replication and functional studies with independent and larger samples will be beneficial to confirm the present findings. Another limitation of our study was that underlying mechanisms explaining the results have not been assessed. An observed statistical association between a genetic marker and a phenotype does not necessarily mean a causal relationship. Further mechanistic investigations are warranted to elucidate the possible mechanisms related to these markers.
In conclusion, the relationship between testosterone levels and muscle fiber size can partly be explained by shared genetic variants. We identified five testosterone-increasing alleles (DOCK3 rs77031559 G, ESR1 rs190930099 G, GLIS3 rs34706136 TG, GRAMD1B rs850294 T, TRAIP rs62260729 C) that were also associated with CSA of fast-twitch muscle fibers, fat-free mass and handgrip strength. Based on these five SNPs, the number of testosterone-increasing alleles was positively associated with testosterone levels and weightlifting performance, as well as participants with ≥ 2 favorable alleles were overrepresented in power athletes. While many more genetic factors undoubtedly remain undiscovered (Ahmetov et al. 2021), these five provide a basis on which future, more comprehensive, genetic assessments might augment systems of identifying and nurturing talent in elite power sports.
Abbreviations
- AR:
-
Androgen receptor
- CSA:
-
Cross-sectional area of muscle fibers
- DOCK3 :
-
Dedicator of cytokinesis 3 gene
- DNA:
-
Deoxyribonucleic acid
- DHT:
-
Dihydrotestosterone
- ESR1 :
-
Estrogen receptor 1 gene
- GWAS:
-
Genome-wide association study
- GLIS3 :
-
GLIS family zinc finger 3 gene
- GRAMD1B :
-
GRAM domain-containing 1B gene
- mTOR:
-
Mammalian target of rapamycin
- PCA:
-
Principal component analysis
- SNPs:
-
Single-nucleotide polymorphisms
- TRAIP:
-
TRAF interacting protein
References
Ackley BD (2014) Wnt-signaling and planar cell polarity genes regulate axon guidance along the anteroposterior axis in C. elegans. Dev Neurobiol 74(8):781–796
Ahmetov II, Donnikov AE, Trofimov DY (2014) Actn3 genotype is associated with testosterone levels of athletes. Biol Sport 31(2):105–108
Ahmetov II, Mozhayskaya IA, Lyubaeva EV, Vinogradova OL, Rogozkin VA (2008) PPARG Gene polymorphism and locomotor activity in humans. Bull Exp Biol Med 146(5):630–632. https://doi.org/10.1007/s10517-009-0364-y
Ahmetov II et al (2020) Is testosterone responsible for athletic success in female athletes? J Sports Med Phys Fitness 60(10):1377–1382
Ahmetov II, Hall ECR, Semenova EA, Pranckevičienė E, Ginevičienė V (2021) Advances in sports genomics. Adv Clin Chem. https://doi.org/10.1016/bs.acc.2021.07.004
Alexander MS et al (2014) MicroRNA-486-dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy-associated symptoms. J Clin Invest 124(6):2651–2667
Basualto-Alarcon C, Jorquera G, Altamirano F, Jaimovich E, Estrada M (2013) Testosterone signals through mTOR and androgen receptor to induce muscle hypertrophy. Med Sci Sports Exerc 45(9):1712–1720
Bermon S, Garnier PY (2017) Serum androgen levels and their relation to performance in track and field: mass spectrometry results from 2127 observations in male and female elite athletes. Br J Sports Med 51(17):1309–1314
Bermon S et al (2014) Serum androgen levels in elite female athletes. J Clin Endocrinol Metab 99(11):4328–4335
Bhasin S et al (2001) Testosterone dose-response relationships in healthy young men. Am J Physiol Endocrinol Metab 281(6):E1172-1181
Bhasin S et al (2005) Older men are as responsive as young men to the anabolic effects of graded doses of testosterone on the skeletal muscle. J Clin Endocrinol Metab 90(2):678–688
Broos S et al (2016) Evidence for ACTN3 as a speed gene in isolated human muscle fibers. PLoS ONE 11(3):e0150594
Brown M, Ning J, Ferreira JA, Bogener JL, Lubahn DB (2009) Estrogen receptor-alpha and -beta and aromatase knockout effects on lower limb muscle mass and contractile function in female mice. Am J Physiol Endocrinol Metab 296(4):E854-861
D’Andrea S, Spaggiari G, Barbonetti A, Santi D (2020) Endogenous transient doping: physical exercise acutely increases testosterone levels-results from a meta-analysis. J Endocrinol Invest 43(10):1349–1371
Dubois V, Laurent M, Boonen S, Vanderschueren D, Claessens F (2012) Androgens and skeletal muscle: cellular and molecular action mechanisms underlying the anabolic actions. Cell Mol Life Sci 69(10):1651–1667
Duncan L, Shen H, Gelaye B, Meijsen J, Ressler K, Feldman M, Peterson R, Domingue B (2019) Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun 10(1):3328
Grishina EE et al (2019) Three DNA polymorphisms previously identified as markers for handgrip strength are associated with strength in weightlifters and muscle fiber hypertrophy. J Strength Cond Res 33(10):2602–2607
Guilherme J, Lancha AH Jr (2020) Total genotype score and athletic status: an exploratory cross-sectional study of a Brazilian athlete cohort. Ann Hum Genet 84(2):141–150
Guilherme J et al (2021a) Are genome-wide association study identified single-nucleotide polymorphisms associated with sprint athletic status? A replication study with 3 different cohorts. Int J Sports Physiol Perform 16(4):489–495
Guilherme JPLF, Shikhova YV, Dondukovskaya RR, Topanova AA, Semenova EA, Astratenkova IV, Ahmetov II (2021b) Androgen receptor gene microsatellite polymorphism is associated with muscle mass and strength in bodybuilders and power athlete status. Ann Hum Biol. https://doi.org/10.1080/03014460.2021.1919204
Helbig KL, Mroske C, Moorthy D, Sajan SA, Velinov M (2017) Biallelic loss-of-function variants in DOCK3 cause muscle hypotonia, ataxia, and intellectual disability. Clin Genet 92(4):430–433
Hong Y et al (2001) Familial resemblance for free androgens and androgen glucuronides in sedentary black and white individuals: the HERITAGE Family Study. Health, risk factors exercise training and genetics. J Endocrinol 170(2):485–492
Horwath O et al (2020) Fiber type-specific hypertrophy and increased capillarization in skeletal muscle following testosterone administration in young women. J Appl Physiol 128(5):1240–1250
Jungmichel S, Sylvestersen KB, Choudhary C, Nguyen S, Mann M, Nielsen ML (2014) Specificity and commonality of the phosphoinositide-binding proteome analyzed by quantitative mass spectrometry. Cell Rep 6(3):578–591
Larsen MC, Lee J, Jorgensen JS, Jefcoate CR (2020) STARD1 functions in mitochondrial cholesterol metabolism and nascent HDL formation. gene expression and molecular mRNA imaging show novel splicing and a 1:1 mitochondrial association. Front Endocrinol (lausanne) 11:559674
Lasevicius T et al (2018) Effects of different intensities of resistance training with equated volume load on muscle strength and hypertrophy. Eur J Sport Sci 18(6):772–780
Lemoine S, Granier P, Tiffoche C, Rannou-Bekono F, Thieulant ML, Delamarche P (2003) Estrogen receptor alpha mRNA in human skeletal muscles. Med Sci Sports Exerc 35(3):439–443
Lenina O, Petrov K, Kovyazina I, Malomouzh A (2019) Enhancement of mouse diaphragm contractility in the presence of antagonists of GABAA and GABAB receptors. Exp Physiol 104(7):1004–1010
Miller R et al (2020) The muscle morphology of elite sprint running. Med Sci Sports Exerc 53(4):804–815
Moreland E et al (2020) Polygenic profile of elite strength athletes. J Strength Cond Res. https://doi.org/10.1519/JSC.0000000000003901
Naito T et al (2019) Movement of accessible plasma membrane cholesterol by the GRAMD1 lipid transfer protein complex. Elife. https://doi.org/10.7554/eLife.51401
Pimenta EM et al (2012) The ACTN3 genotype in soccer players in response to acute eccentric training. Eur J Appl Physiol 112(4):1495–1503
Ruth KS et al (2020) Using human genetics to understand the disease impacts of testosterone in men and women. Nat Med 26(2):252–258
Seaborne RA et al (2019) UBR5 is a novel E3 ubiquitin ligase involved in skeletal muscle hypertrophy and recovery from atrophy. J Physiol 597(14):3727–3749
Sinha-Hikim I et al (2002) Testosterone-induced increase in muscle size in healthy young men is associated with muscle fiber hypertrophy. Am J Physiol Endocrinol Metab 283(1):E154-164
Thalacker-Mercer A, Stec M, Cui X, Cross J, Windham S, Bamman M (2013) Cluster analysis reveals differential transcript profiles associated with resistance training-induced human skeletal muscle hypertrophy. Physiol Genomics 45(12):499–507
Travison TG et al (2014) The heritability of circulating testosterone, oestradiol, oestrone and sex hormone binding globulin concentrations in men: the Framingham Heart Study. Clin Endocrinol (oxf) 80(2):277–282
Vissing K, Schjerling P (2014) Simplified data access on human skeletal muscle transcriptome responses to differentiated exercise. Sci Data 1:140041
Wood RI, Stanton SJ (2012) Testosterone and sport: current perspectives. Horm Behav 61(1):147–155
Wood AR et al (2014) Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 46(11):1173–1186
Zeng F, Zhao H, Liao J (2017) Androgen interacts with exercise through the mTOR pathway to induce skeletal muscle hypertrophy. Biol Sport 34(4):313–321
Zhang JL, Zhang CL, Zhou BG, Lei BY, Zhang B, Yang HT (2021) Association study of the functional variants of the GLIS3 gene with risk of knee osteoarthritis. Clin Rheumatol 40(3):1039–1046
Acknowledgements
The study was supported in part by grant from the Russian Science Foundation (Grant No. 21-15-00362: “Study of molecular and genetic mechanisms of morphofunctional changes in human muscle fibers during high-intensity physical training”). The authors would like to thank Evgeny A. Lysenko, Tatiana F. Vepkhvadze, Egor M. Lednev and Daniil V. Popov for their help with determination of muscle fiber composition.
Author information
Authors and Affiliations
Contributions
IIA and EVG conceived and designed the study. JPLFG, EAS, OVB, AKL, EM, EVG and IIA acquired, analyzed, and interpreted data. JPLFG and IIA drafted the manuscript and all authors reviewed and revised it critically for important intellectual content. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Additional information
Communicated by Philip D Chilibeck.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guilherme, J.P.L.F., Semenova, E.A., Borisov, O.V. et al. Genomic predictors of testosterone levels are associated with muscle fiber size and strength. Eur J Appl Physiol 122, 415–423 (2022). https://doi.org/10.1007/s00421-021-04851-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00421-021-04851-w