Abstract
The genetic polymorphisms of 21 autosomal short tandem repeat (STR) loci included in the GlobalFiler™ PCR Amplification Kit were evaluated in 3032 unrelated individuals Altay Han of Xinjiang, northwest China. All of the loci reached the Hardy-Weinberg equilibrium. These loci were examined to determine allele frequencies and forensic statistical parameters. SE33 showed the greatest power of discrimination in Altay Han population, whereas TPOX showed the lowest. The combined discrimination power and probability of excluding paternity of the 21 autosomal STR loci were 0.999999999999999999999999889838 and 0.999999996664704, respectively. Both pairwise genetic distance and phylogenetic methods indicated that the Altay Han had the closest genetic relationship with the Han origin and Hui populations. The present results revealed that the GlobalFiler system had a high level of polymorphism in Altay Han population and hence could be a powerful tool for forensic application and population genetic study.
Similar content being viewed by others
References
Wang DY, Gopinath S, Lagace RE, Norona W, Hennessy LK, Short ML, Mulero JJ (2015) Developmental validation of the GlobalFiler® Express PCR Amplification Kit: a 6-dye multiplex assay for the direct amplification of reference samples. Forensic Sci Int Genet 19:148–155
Hennessy LK, Mehendale N, Chear K, Jovanovich S, Williams S, Park C, Gangano S (2014) Developmental validation of the GlobalFiler® express kit, a 24-marker STR assay, on the RapidHIT® System. Forensic Sci Int Genet 13:247–258
Hammond HA, Jin L, Zhong Y, Caskey CT, Chakraborty R (1994) Evaluation of 13 short tandem repeat loci for use in personal identification applications. Am J Hum Genet 55:175–189
Butler JM, Hill CR (2012) Biology and genetics of new autosomal STR loci useful for forensic DNA analysis. Forensic Sci Rev 24:15–26
Hares DR (2015) Selection and implementation of expanded CODIS core loci in the United States. Forensic Sci Int Genet 17:33–34
Hares DR (2012) Expanding the CODIS core loci in the United States. Forensic Sci Int Genet 6:e52–e54
Walsh PS, Metzger DA, Higuchi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques 10:506–513
Bar W, Brinkmann B, Budowle B, Carracedo A, Gill P, Lincoln P, MayrW OB (1997) DNA recommendations. Further report of the DNA commission of the ISFH regarding the use of short tandem repeat systems. International Society for Forensic Haemogenetics. Int J Legal Med 110:175–176
Lincoln PJ (1997) DNA recommendations—further report of the DNA commission of the ISFH regarding the use of short tandem repeat systems. Forensic Sci Int 87:181–184
Zhao F, Wu XY, Cai GQ, Xu CC (2003) The application ofmodified-Powerstates software in forensic biostatistics (in Chinese). Chin J Forensic Med 18:297–298
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Weir B, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370
Weir BS, Hill WG (2002) Estimating F-statistics. Annu Rev Genet 36:721–750
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Ramos-González B, Aguilar-Velázquez JA, Chávez-Briones MDL, Delgado-Chavarría JR, Alfaro-Lopez E, Rangel-Villalobos H (2016) Population data of 24 STRs in Mexican-Mestizo population from Monterrey, Nuevo Leon (Northeast, Mexico) based on Powerplex® Fusion and GlobalFiler® kits. Forensic Sci Int Genet 21:e15–e17
Guerreiro S, Ribeiro T, Porto MJ, Carneiro De Sousa MJ, Dario P (2017) Characterization of GlobalFiler loci in Angolan and Guinean populations inhabiting Southern Portugal. Int J Legal Med 131:365–368
Ng J, Oldt RF, McCulloh KL, Weise JA, Viray J, Budowle B, Smith DG, Kanthaswamy S (2016) Native American population data based on the Globalfiler® autosomal STR loci. Forensic Sci Int Genet 24:e12–e13
Carracedo A, Butler JM, Gusmao L, Linacre A, Parson W, Roewer L, Schneider PM (2014) Update of the guidelines for the publication of genetic population data. Forensic Sci Int Genet 10:1–2
Acknowledgements
This study was supported by the Department of Education, Guangdong Government under the Top-tier University Development Scheme for Research and Control of Infectious Diseases (No. 2015064), Natural Science Foundation of Shandong Province (No. ZR2014HQ018). National Natural Science Foundation Council of China (No. 81671874). Project of Shandong Province Higher Educational Science and Technology Program (No. J17KA240), China Postdoctoral Science Foundation (No. 2017M612701).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Fig. S1
The locations of Altay and other Han populations in China a: China, b: Xinjiang (DOCX 2421 kb).
Fig. S2
Multidimensional scaling (MDS) plot based on pairwise Fst values between Altay Han and other populations (DOCX 29 kb).
Fig. S3
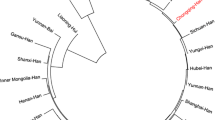
The neighbor-joining trees constructed with pairwise Fst (DOCX 17 kb).
Table S1
Allele frequencies and forensic parameters of 21 autosomal STRs in Altay Han population living in Xinjiang, China (n=3,032) (XLSX 19.4 kb).
Table S2
P-values of LD test between 21 autosomal STR loci in Altay Han population living in Xinjiang, China (n=3,032) (XLSX 12 kb).
Table S3
P-values of population differentiation exact test between Altay Han and the reference populations (XLSX 14 kb).
Table S4
Fst values between pairwise populations in the present study by using 21 autosomal STR loci (XLSX 10 kb).
Rights and permissions
About this article
Cite this article
Li, X., Li, L., Wang, Q. et al. Population genetic analysis of the Globalfiler STR loci in 3032 individuals from the Altay Han population of Xinjiang in northwest China. Int J Legal Med 132, 141–143 (2018). https://doi.org/10.1007/s00414-017-1641-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-017-1641-3