Abstract
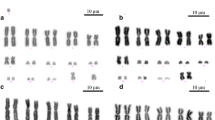
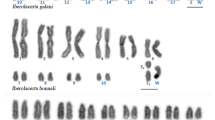
The sand lizard (Lacerta agilis, Lacertidae) has a chromosome number of 2n = 38, with 17 pairs of acrocentric chromosomes, one pair of microchromosomes, a large acrocentric Z chromosome, and a micro-W chromosome. To investigate the process of karyotype evolution in L. agilis, we performed chromosome banding and fluorescent in situ hybridization for gene mapping and constructed a cytogenetic map with 86 functional genes. Chromosome banding revealed that the Z chromosome is the fifth largest chromosome. The cytogenetic map revealed homology of the L. agilis Z chromosome with chicken chromosomes 6 and 9. Comparison of the L. agilis cytogenetic map with those of four Toxicofera species with many microchromosomes (Elaphe quadrivirgata, Varanus salvator macromaculatus, Leiolepis reevesii rubritaeniata, and Anolis carolinensis) showed highly conserved linkage homology of L. agilis chromosomes (LAG) 1, 2, 3, 4, 5(Z), 7, 8, 9, and 10 with macrochromosomes and/or macrochromosome segments of the four Toxicofera species. Most of the genes located on the microchromosomes of Toxicofera were localized to LAG6, small acrocentric chromosomes (LAG11–18), and a microchromosome (LAG19) in L. agilis. These results suggest that the L. agilis karyotype resulted from frequent fusions of microchromosomes, which occurred in the ancestral karyotype of Toxicofera and led to the disappearance of microchromosomes and the appearance of many small macrochromosomes.







Similar content being viewed by others
References
Alföldi J, Di Palma F, Grabherr M et al (2011) The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477:587–591. doi:10.1038/nature10390
Ciofi C, Swingland IR (1997) Environmental sex determination in reptiles. Appl Anim Behav Sci 51:251–265
Cree A, Thompson MB, Daugherty CH (1995) Tuatara sex determination. Nature 375:543. doi:10.1038/375543a0
De Smet WHO (1981) Description of the orcein stained karyotypes of 27 lizard species (Lacertilia, Reptilia) belonging to the families Teiidae, Scincidae, Lacertidae, Cordylidae and Varanidae (Autarchoglossa). Acta Zool Pathol Antverpiensia 76:73–118
Ezaz T, Sarre SD, O’Meally D, Marshall Graves JA, Georges A (2010) Sex chromosome evolution in lizards: independent origins and rapid transitions. Cytogenet Genome Res 127:249–260. doi:10.1159/000300507
Ezaz T, Azad B, O’Meally D et al (2013) Sequence and gene content of a large fragment of a lizard sex chromosome and evaluation of candidate sex differentiating gene R-spondin 1. BMC Genomics 14:899. doi:10.1186/1471-2164-14-899
Gamble T, Geneva AJ, Glor RE, Zarkower D (2013) Anolis sex chromosomes are derived from a single ancestral pair. Evolution. doi:10.1111/evo.12328
Head G, May RM, Pendleton L (1987) Environmental determination of sex in the reptiles. Nature 329:198–199. doi:10.1038/329198a0
International Chicken Genome Sequencing Consortium (ICGSC) (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716. doi:10.1038/nature03154
Kawagoshi T, Uno Y, Matsubara K, Matsuda Y, Nishida C (2009) The ZW micro-sex chromosomes of the Chinese soft-shelled turtle (Pelodiscus sinensis, Trionychidae, Testudines) have the same origin as chicken chromosome 15. Cytogenet Genome Res 125:125–131. doi:10.1159/000227837
Kawagoshi T, Nishida C, Matsuda Y (2012) The origin and differentiation process of X and Y chromosomes of the black marsh turtle (Siebenrockiella crassicollis, Geoemydidae, Testudines). Chromosome Res 20:95–110. doi:10.1007/s10577-011-9267-7
Kawai A, Ishijima J, Nishida C, Kosaka A, Ota H, Kohno S, Matsuda Y (2009) The ZW sex chromosomes of Gekko hokouensis (Gekkonidae, Squamata) represent highly conserved homology with those of avian species. Chromosoma 118:43–51. doi:10.1007/s00412-008-0176-2
Lang JW, Andrews HV (1994) Temperature-dependent sex determination in crocodilians. J Exp Zool 270:28–44. doi:10.1002/jez.1402700105
Leaché AD, Sites JW Jr (2009) Chromosome evolution and diversification in north American spiny lizards (Genus Sceloporus). Cytogenet Genome Res 127:166–181. doi:10.1159/000293285
Matsubara K, Tarui H, Toriba M et al (2006) Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad Sci U S A 103:18190–18195. doi:10.1073/pnas.0605274103
Matsubara K, Kuraku S, Tarui H et al (2012) Intra-genomic GC heterogeneity in sauropsids: evolutionary insights from cDNA mapping and GC3 profiling in snake. BMC Genomics 13:604. doi:10.1186/1471-2164-13-604
Matsuda Y, Chapman VM (1995) Application of fluorescence in situ hybridization in genome analysis of the mouse. Electrophoresis 16:261–272
Matsuda Y, Nishida-Umehara C, Tarui H et al (2005) Highly conserved linkage homology between birds and turtles: bird and turtle chromosomes are precise counterparts of each other. Chromosome Res 13:601–615. doi:10.1007/s10577-005-0986-5
Nishida-Umehara C, Tsuda Y, Ishijima J, Ando J, Fujiwara A et al (2007) The molecular basis of chromosome orthologies and sex chromosomal differentiation in palaeognathous birds. Chromosome Res 15:721–734. doi:10.1007/s10577-007-1157-7
Odierna G, Olmo E, Cobror O (1987) Taxonomic implications of NOR-localization in lacertid lizards. Amphibia-Reptilia 87:373–382
Odierna G, Kupriyanova L, Capriglione T, Olmo E (1993) Further data on sex chromosomes of Lacertidae and a hypothesis on their evolutionary trend. Amphibia-Reptilia 14:1–11
Odierna G, Olmo E, Caputo V, Capriglione T, in den Bosch HAJ (1995) Karyological affinity between Lacerta fraasii Lehrs, 1910 and Lacerta parva Boulenger, 1887. Amphibia-Reptilia 16:293–297
Olmo E, Signorino G (2005) Chromorep: a reptile chromosomes database. Internet references. Retrieved from: http://chromorep.univpm.it. 6/04/2013
Olmo E, Odierna G, Capriglione T (1987) Evolution of sex-chromosomes in lacertid lizards. Chromosoma 96:33–38
Olmo E, Odierna G, Capriglione T, Caputo V (1991) A karyological approach to the systematic of Lacertidae (Reptilia, Sauria). Rev Esp Herpetol 6:81–90
Pokorná M, Kratochvíl L, Kejnovský E (2011) Microsatellite distribution on sex chromosomes at different stages of heteromorphism and heterochromatinization in two lizard species (Squamata: Eublepharidae: Coleonyx elegans and Lacertidae: Eremias velox). BMC Genet 12:90. doi:10.1186/1471-2156-12-90
Pyron RA, Burbrink FT, Wiens JJ (2013) A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol 13:93. doi:10.1186/1471-2148-13-93
Shedlock AM, Edwards SV (2009) Amniota. In: Hedges SB, Kumar S (eds) The timetree of life. Oxford University Press, New York, pp 375–379
Srikulnath K, Matsubara K, Uno Y et al (2009a) Karyological characterization of the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Squamata) by molecular cytogenetic approach. Cytogenet Genome Res 125:213–223. doi:10.1159/000230005
Srikulnath K, Nishida C, Matsubara K et al (2009b) Karyotypic evolution in squamate reptiles: comparative gene mapping revealed highly conserved linkage homology between the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Lacertilia) and the Japanese four-striped rat snake (Elaphe quadrivirgata, Colubridae, Serpentes). Chromosome Res 17:975–986. doi:10.1007/s10577-009-9101-7
Srikulnath K, Uno Y, Nishida C, Matsuda Y (2013) Karyotype evolution in monitor lizards: cross-species chromosome mapping of cDNA reveals highly conserved synteny and gene order in the Toxicofera clade. Chromosome Res 21:805–819. doi:10.1007/s10577-013-9398-0
Uetz P (2014) The TIGR reptile database. The EMBL reptile database. Retrieved from: http://www.reptile-database.org/
Uno Y, Nishida C, Tarui H et al (2012) Inference of the protokaryotypes of amniotes and tetrapods and the evolutionary processes of microchromosomes from comparative gene mapping. PLoS ONE 7:e53027. doi:10.1371/journal.pone.0053027
Vidal N, Hedges SB (2005) The phylogeny of squamate reptiles (lizards, snakes, and amphisbaenians) inferred from nine nuclear protein-coding genes. C R Biol 328:1000–1008. doi:10.1016/j.crvi.2005.10.001
Acknowledgments
This work was financially supported by Grants-in-Aid for Scientific Research on Innovative Areas (No. 23113004) and Scientific Research (B) (No. 22370081) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
(XLS 36 kb)
Rights and permissions
About this article
Cite this article
Srikulnath, K., Matsubara, K., Uno, Y. et al. Identification of the linkage group of the Z sex chromosomes of the sand lizard (Lacerta agilis, Lacertidae) and elucidation of karyotype evolution in lacertid lizards. Chromosoma 123, 563–575 (2014). https://doi.org/10.1007/s00412-014-0467-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-014-0467-8