Abstract
Purpose
To investigate Src-like adaptor 2 gene (SLA2) expression in head and neck squamous cell carcinoma (HNSCC), its potential prognostic value, and its effect on immune cell infiltration.
Methods
Through a variety of bioinformatics analyses, we extracted and analyzed data sets from the Cancer Genome Atlas (TCGA), Tumor Immune Estimation Resource (TIMER), and Gene Expression Profile Interaction Analysis (GEPIA) to analyze the correlation between SLA2 and the prognosis, immune checkpoint, tumor microenvironment (TME) and immune cell infiltration of HNSCC, and to explore its potential oncogenic mechanism. To further explore the potential role of SLA2 in HNSCC by Gene ontology (GO) functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.
Results
SLA2 messenger ribonucleic acid (mRNA) levels were increased in HNSCC tumor tissues compared with normal tissues. In addition, we found that SLA2 may be an independent prognostic factor for HNSCC, and high SLA2 expression is associated with favorable prognosis in HNSCC. SLA2 expression was positively correlated with B cells, cluster of differentiation 8-positive T cells (CD8 + T cells), cluster of differentiation 4-positive T cells (CD4 + T cells), macrophages, neutrophil and dendritic cells infiltration. SLA2 has also been shown to co-express immune-related genes and immune checkpoints. Significant GO term analysis by Gene Set Enrichment Analysis (GSEA) indicated that genes correlated with SLA2 were located mainly in the side of membrane, receptor complex, secretory granule membrane, endocytic vesicle, membrane region, and endosome membrane, where they were involved in leukocyte cell–cell adhesion, response to interferon-gamma, and regulation of immune effector process. These related genes also served as antigen binding, cytokine receptor activity, phosphatidylinositol 3-kinase activity, peptide receptor activity, Src homology domain 3 (SH3) domain binding, and cytokine receptor binding. KEGG pathway analysis demonstrated that these genes related to SLA2 were mainly enriched in signal pathways, such as hematopoietic cell lineage, cell adhesion molecules (CAMs), natural killer cell mediated cytotoxicity, measles, and chemokine signaling pathway.
Conclusions
SLA2 is increased in HNSCC, and high SLA2 expression is associated with favorable prognosis. SLA2 may affect tumor development by regulating tumor infiltrating cells in TME. SLA2 may be a potential target for immunotherapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Head and neck squamous cell carcinoma (HNSCC), which develops from the mucosal epithelium of the mouth, pharynx and larynx, is the most common malignant tumor of the head and neck. It is generally found in the late stage, with a high degree of malignancy and a poor prognosis. At present, although tumor resection combined with chemotherapy and radiotherapy has improved the prognosis of patients to a certain extent, advanced cancer is still prone to metastasis and deterioration. Therefore, it is necessary to explore more effective treatment methods in advanced HNSCC patients, such as targeted therapy. However, in the traditional research of targeted therapy for malignant tumors, the molecular pathways seem to be too complex for researchers to fully understand. So far, no effective biomarkers have been found [1].
Therefore, in this study, a variety of bioinformatics methods were used to explore the potential oncogenic mechanism of Src-like adaptor 2 gene (SLA2) in HNSCC, and the Cancer Genome Atlas (TCGA), Tumor Immune Estimation Resource (TIMER), Gene Expression Profile Interaction Analysis (GEPIA) and LinkedOmics data sets were extracted and analyzed. The correlation between SLA2 and tumor immune cell infiltration, immune checkpoint, immune-related genes and immune checkpoint blockade (ICB) was detected, Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG), pathway enrichment analyses were applied to investigate the potential functions of SLA2. These results provide new insights into the function of SLA2 and new targets for the diagnosis and prognosis of HNSCC.
SLA2 is an adaptor protein that negatively regulates T-cell receptor (TCR) signaling. Inhibition of T cell antigen receptor induces activation of activated T cell nuclear factor. It may act by linking zeta chain of T cell receptor-associated protein kinase 70 (ZAP70) and other signaling proteins to Casitas B-lineage lymphoma proto-oncogene (CBL), resulting in CBL-dependent degradation of signaling proteins. It is involved in the development of various tumors, such as non-small cell lung cancer, breast cancer, and pancreatic duct adenocarcinoma [2,3,4]. However, whether SLA2 affects the prognosis of HNSCC has not been investigated.
In recent years, immunotherapy has been applied to the treatment of HNSCC patients, changing the treatment of HNSCC [5]. It is well known that tumor-infiltrating immune cells (TIICs) affect the immune system, deal with abnormal biological behaviors in a complex way, and play an important role in response to immunotherapy [6]. In addition, genes related to immune components can also affect immune function [7]. Moreover, cytokine and immune checkpoint blockade therapy has become a therapeutic strategy for various types of cancer, but no study has investigated whether SLA2 overexpression affects the tumor immune microenvironment of HNSCC.
Methods
Data source and analysis of differential expressions
Data acquisition, preprocessing, and ethics statement Pan-cancer ribonucleic acid sequencing (RNA-seq) data were obtained from UCSC XENA database (https:// xenabrowser.net/ datap ages/) [8]. Level 3 RNA RNA-seq expression data and clinical data for HNSCC (502 HNSCC tissues vs. 44 normal adjacent cancer tissues) were downloaded from TCGA portal (https:// portal. gdc. cancer. gov/) [9]. Before analysis, transcripts per million reads (TPM) RNA-seq data were log transformed (Log2(TPM + 1)). We use TCGA to analyze the SLA2 expression difference between HNSCC and normal head and neck tissues. As TCGA are open publicly available databases, the data collection from the databases was compliant with all applicable laws, regulations, and policies for the protection of human subjects, and all written informed consents were obtained from all subjects involved.
Exploring the association of survival with SLA2 in HNSCC
We performed univariate and multivariate Cox regression analyses to determine the appropriate terms for building nomograms. A forest was applied to display the p value, hazard ratio (HR), and 95% confidence intervals (95% CI) for each variable using the “forestplot” R package through R software. A nomogram was created according to the results of the multivariate Cox proportional hazards analysis to predict the total recurrence rate in 1, 3, and 5 years. The “rms” R package was used to evaluate the risk of recurrence for individual patients by the points associated with each risk factor. We extracted survival information for each sample in TCGA. Indicators such as overall survival (OS) and disease-specific survival (DSS) were used to elucidate the relationship betweens SLA2 and the prognosis of HNSCC patients. Kaplan–Meier (KM) and log-rank tests were used for the survival analysis of HNSCC (p < 0.05), and survival curves were analyzed by the “survminer” and “survivor” R packages.
GEPIA analysis
GEPIA is a Newly developed interactive web server. Its web site is http://gepia.cancer-pku.cn/index.html [10]. This database provides online analysis based on data from TCGA and the Genotype-Tissue Expression (GTeX) projects. In our study, we used the given TCGA expression data set to assess the difference of SLA2 between HNSCC and normal head and neck tissues. In addition, we used GEPIA to analyze the prognostic significance of SLA2 mRNA expression in HNSCC and draw the survival curve with log rank p value.
Immune infiltration analysis
Single-sample Gene Set Enrichment Analysis (ssGSEA) was used to assess the infiltration of individual immune cell populations, e.g., T helper 2 cells (Th2 cells), cluster of differentiation 8 T cells (CD8 T cells), natural killer CD56dim cells (NK CD56dim cells), and others. The analysis was performed using Gene Set Variation Analysis (GSVA) package [11, 12]. The TIMER (https://cistrome.shinyapps.io/timer/) database was applied to explore the correlations between HNSCC and TIICs (Li et al. 2017) [13]. GSVA package of R was used to analyze the correlations between SLA2 expression levels and the infiltrations of T cells, B cells, regulatory T cell (TReg), CD8 T cells, dendritic cell (DC), activated dendritic cell (aDC), Cytotoxic cells, effective memory T cell (Tem), Eosinophils, and NK CD56dim cells.
Immune-checkpoint analysis
Sialic acid binding Ig-like lectin 15 (SIGLEC15), T cell immunoreceptor with Ig and ITIM domains (TIGIT), cytotoxic T lymphocyte-associated antigen-4 (CTLA4), cluster of differentiation 274 (CD274), hepatitis A virus cellular receptor 2 (HAVCR2), lymphocyte-activation gene 3 (LAG3), programmed cell death 1 (PDCD1), and programmed cell death 1 ligand 2 (PDCD1LG2) were selected to be immune-checkpoint-relevant transcripts, and the expression data of these eight genes were extracted. R package “ggplot2” “pheatmap” and “immuneeconv” were used to assess the expression of the immune-checkpoints and co-expression of SLA2 with these immune-checkpoints. Potential immune checkpoint blockade response was predicted with the TIDE algorithm (Jiang et al. 2018) [14].
Spearman correlation analysis of IC50 score and SLA2 expression
RNA-sequencing expression profiles and corresponding clinical information for HNSCC were downloaded from the TCGA data set (https://portal.gdc.com). Predicting the chemotherapeutic response for each sample based on the largest publicly available pharmacogenomics database [the Genomics of Drug Sensitivity in Cancer (GDSC), https://www.cancerrxgene.org/] [15,16,17]. The prediction process was implemented by R package “pRRophetic”. The samples' half-maximal inhibitory concentration (IC50) was estimated by ridge regression. All the parameters were set as the default values. Using the batch effect of the combat and tissue type of all tissue, the duplicate gene expression was summarized as the mean value using the batch effect of the combat and tissue type of all tissue. Predicting the relative IC50 of cisplatin and methotrexate in patients with HNSCC.
LinkedOmics analysis
LinkedOmics is a public website on http://www.linkedomics.org/login.php, which contains 32 TCGA cancer types of multiple omics data, including federation generated from the Clinical Proteomics Tumor Analysis Consortium [18]. We applied this online tool to identify and analyze SLA2 co-expression genes in head and neck cancer cohort from TCGA, which contains 502 samples. The data from Linkfinder are signed and sorted; after that, GO analyses, including molecular function (MF), biological process (BP), cellular component (CC), and KEGG analysis, are performed by the GSEA method.
Statistical analysis
Gene expression data were normalized by log2-transformation. Statistical analyses were conducted using R software (version4.0.2). The analysis of SLA2 expression in TIMER and TCGA databases showed p value. The survival curve generated by GEPIA analysis showed p value. The Cox proportional hazards model, KM analyses, and log-rank test were conducted for all survival analyses. Spearman’s test or Pearson’s test was applied to analyze the correlation between two variables; p value < 0.05 was considered significant. R software (version 4.0.2) was used for statistical analysis.
Results
SLA2 expression in pan-cancer and HNSCC patients
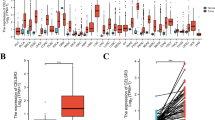
Using the data from TCGA, we found that SLA2 expression was significantly elevated in 19 of 33 cancer types, including kidney renal papillary cell carcinoma (KIRP), kidney renal clear cell carcinoma (KIRC), stomach adenocarcinoma (STAD), among others (Fig. 1A). Similarly, SLA2 expression was significantly increased in HNSCC tissues compared with normal samples (Fig. 1B). It has the same result as in Fig. 1A, with a p value less than 0.001. For the 43 paired samples, SLA2 expression was also significantly higher in cancer tissues than in matched normal adjacent tissue samples (Fig. 1C).
SLA2 expression in pan-cancer and HNSCC patients. A SLA2 expression levels in different tumor types. B SLA2 expression is significantly higher in HNSCC tissues than in normal tissues. ***p < 0.001. C SLA2 expression was also significantly higher in 43 HNSCC tissues than in matched normal adjacent tissue samples. *p < 0.05
The expression of SLA2 Is associated with patients’ survival in HNSCC
The survival data of SLA2 expression in HNSCC patients were analyzed. KM survival curves were used to evaluate the relationship between the SLA2 expression and survival outcomes. The cut-off value of the high and low SLA2 expression group was set as the median. The results showed that patients with higher SLA2 mRNA expression had longer OS (p < 0.019) (Fig. 2A), DSS (p < 0.0277) (Fig. 2B). Consequently, the expression of SLA2 mRNA in HNSCC is associated with survival. The result from GEPIA is also consistent with our results (Fig. 2C).
SLA2 expression level has potential diagnostic and prognostic values for patients with HNSCC. A KM survival curves for OS showed that patients with higher SLA2 mRNA expression had longer OS (p < 0.019). B KM survival curves for DSS showed that patients with higher SLA2 mRNA expression had longer DSS (p < 0.0277). C K–M survival curves for OS constructed from GEPIA
Prognostic potential of SLA2 in HNSCC
We then performed univariate and multivariate Cox regression analyses using several clinical features and SLA2 expression level. Furthermore, univariate and multivariate Cox regression analyses illustrated that SLA2 expression (p < 0.01), pN-stage (p < 0.05), and pM-stage (p < 0.01) were important independent factors to the prognosis of HNSCC (Fig. 3A, B). We further constructed a nomogram that combined only three independent prognostic factors (including SLA2, pN-stage, and pM-stage) to provide a quantitative guideline for clinicians to predict the probability of 1-, 3-, and 5-year OS in HNSCC patients (Fig. 3C). Each patient is given a total score through adding each prognostic parameter point, with a higher total score meaning a worse outcome for that patient. In addition, the calibration curves showed that the nomogram performed well in estimating 1-, 3-, and 5-year OS (Fig. 3D). Therefore, SLA2 may be a potential diagnostic marker for HNSCC.
Prognostic efficacy of SLA2 expression levels in patients with HNSCC. A Univariate Cox regression analyses displayed using forest plots showed that SLA2 expression, pN-stage, and pM-stage were important factors to the prognosis of HNSCC. B Multivariate Cox regression analyses displayed using forest plots showed that SLA2 expression, pN-stage, and pM-stage were important independent factors to the prognosis of HNSCC. C Nomogram for predicting clinical outcomes associated with SLA2 expression in HNSCC patients. D Calibration curves showed that the nomogram performed well in estimating 1-, 3-, and 5-year OS
Association between SLA2 expression and immune cell infiltration and immune molecule expression levels
To understand the effects of SLA2 expression on tumor microenvironment, immune infiltration analysis was performed using ssGSEA method. The correlations between enrichments of immune cell in Head and neck tissues and SLA2 expression levels were calculated using Spearman correlation analyses. As shown in Fig. 4A, SLA2 expression levels were positively correlated with numerous types of immune cells, e.g., T cells, Cytotoxic cells, TReg, aDC, NK CD56dim cells, follicular helper T cell (TFH), CD8 T cells, T helper cells, T helper 1 cells (Th1 cells), B cells, DC, plasmacytoid dendritic cells (pDC), Tem cells, Macrophages, Th2 cells, Eosinophils, interdigitating dendritic cell (iDC), Th17 cells, NK cells, central memory T cells (Tcm), Mast cells, and Neutrophils, all of which play an important role in anti-tumor immunity. Next, we further used the CIBERSORT algorithm to investigate the tumor-infiltrating immune cells, we found that the expression of SLA2 was positively correlated with the infiltration of B cells, cluster of differentiation 8-positive T cells (CD8 + T cells), cluster of differentiation 4-positive T cells (CD4 + T cells), Macrophages cells, Neutrophils cells, and Dendritic cells (Fig. 4B). Similarly, the infiltrations of other immune cells in Head and neck tissues are observed, GSVA package of R was used to analyze the correlations between SLA2 expression levels and the infiltrations of T cells, B cells, TReg, CD8 T cells, DC, aDC, Cytotoxic cells, Tem, Eosinophils, and NK CD56dim cells. As shown in Fig. 4C, SLA2 expression levels were significantly positively correlated with the infiltrations of those immune cells (p < 0.01).
Relationship between SLA2 expression and immune cell infiltration in the tumor microenvironment. A SLA2 expression levels were positively correlated with numerous types of 24 immune cells. B Expression of SLA2 was positively correlated with the infiltration of (B), CD8 + , CD4 + , Macrophages, Neutrophils and dendritic cells determined using TIMER. C SLA2 expression levels were significantly positively correlated with T cells, B cells, TReg, CD8 T cells, DC, aDC, Cytotoxic cells, Tem, Eosinophils, and NK CD56dim cells enrichment levels (p < 0.01)
We further performed gene co-expression analysis to evaluate the mechanisms that SLA2 was correlated with the infiltration of immune cells in HNSCC. Major histocompatibility complex gene (MHC genes), immunosuppressive, immune activation genes, chemokine and chemokine receptor-related genes were assessed. The results showed that SLA2 positively co-expressed with all listed MHC genes (Fig. 5A). Notably, almost all immunosuppressive genes positively co-expressed with SLA2 (Fig. 5B). SLA2 had a positive relationship with most of the immune activation genes, such as genes that encoding cluster of differentiation 48 T cells (CD48), cluster of differentiation 28 T cells (CD28), Interleukin-6 (IL6), C–X–C motif chemokine receptor 4 (CXCR4) (Fig. 5C). Similarly, SLA2 had a positive correlation with some chemokine and chemokine receptors (Fig. 5D, E). The expression of immune-checkpoints, including CD274, CTLA4, HAVCR2, LAG3, PDCD1, PDCD1LG2, TIGIT, and SIGLEC15 (Fig. 6A), were further investigated in the World Health Organization (WHO) grade II and III of HNSCC. The results illustrated that LAG3 (p < 3.35e−05), PDCD1 (p < 1.06e−04), CTLA4 (p < 2.55e−03), HAVCR2 (p < 1.72e−03), TIGIT (p < 1.22e−03), and SIGLEC15 (p < 9.39e−02) were upregulated in WHO grade III compared with WHO grade II of HNSCC (Fig. 6B). In addition, we revealed that HNSCC patients in WHO grade III had a better response to immune checkpoint blockade compared with HNSCC patients in WHO grade I (Fig. 6C). Moreover, the results demonstrated that SLA2 positively co-expressed with these immune-checkpoints (Table 1), indicating that SLA2 may be a potential immunotherapy target.
Co-expression of SLA2 with immune-related genes in HNSCC. A SLA2 positively co-expressed with all listed MHC genes. B SLA2 positively co-expressed with almost all immunosuppressive genes. C SLA2 had a positive relationship with most of the immune activation genes. D SLA2 had a positive correlation with some chemokine. E SLA2 had a positive correlation with some chemokine receptors
Expression of immune-checkpoints in HNSCC. A, B Expression of immune-checkpoints were upregulated in WHO grade III compared with WHO grade II of HNSCC. C HNSCC patients in WHO grade III had a better response to immune checkpoint blockade compared with HNSCC patients in WHO grade I. D, E IC50 of cisplatin and methotrexate was negatively correlated with SLA2 expression
Spearman correlation analysis of IC50 score and SLA2 expression
RNA-sequencing expression profiles and corresponding clinical information for HNSCC were downloaded from the TCGA data set (https://portal.gdc.com). The results showed that IC50 of cisplatin and methotrexate was negatively correlated with SLA2 expression (Fig. 6D, E).
Enrichment analysis of SLA2 neighborhood genes in HNSCC
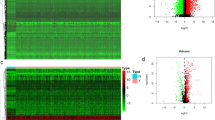
To study the functional network of SLA2 neighborhood genes in HNSCC, we first identified SLA2 neighborhood genes with LinkedOmics. The results are shown in a volcanic plot (Fig. 7A). In addition, the top 50 positively and negatively related genes were shown in the heat map, respectively (Fig. 7B, C). Significant GO term analysis by GSEA indicated that genes correlated with SLA2 were located mainly in the side of membrane, receptor complex, secretory granule membrane, endocytic vesicle, membrane region, and endosome membrane, where they were involved in leukocyte cell–cell adhesion, response to interferon-gamma, and regulation of immune effector process. These related genes also served as antigen binding, cytokine receptor activity, phosphatidylinositol 3-kinase activity, peptide receptor activity, Src homology domain 3 (SH3) domain binding, and cytokine receptor binding (Fig. 7D–F). KEGG pathway analysis demonstrated that these genes related to SLA2 were mainly enriched in signal pathways, such as hematopoietic cell lineage, cell adhesion molecules (CAMs), natural killer cell mediated cytotoxicity, measles, and chemokine signaling pathway (Fig. 7G).
Genes differentially expressed in association with SLA2 in HNSCC, and enrichment analysis of the genes altered in the SLA2 neighborhood in HNSCC. A Volcano plot. B Heat maps of the top 50 genes positively correlated with SLA2. C Heat maps of the top 50 genes negatively correlated with SLA2. D Biological processes. E Cellular components. F Molecular functions. G KEGG pathway analysis
Discussion
HNSCC is the sixth most prevalent cancer in the world. It originates from different parts of the head and neck region, including the oral cavity, nasal cavity, nasopharynx, larynx, hypopharynx, and oropharynx. Squamous cell carcinoma (SCC) accounts for 90% of head and neck malignancies [19]. Due to the characteristics of delayed early diagnosis and high recurrence rate of HNSCC, about 50% of all patients will eventually have local or regional recurrence. Despite the progress in treatment, the 5-year survival rate is still very low, and there are still some patients with poor treatment effect and poor prognosis [20]. Therefore, it is of great significance to explore new genes related to the occurrence and development of HNSCC, and to find markers with higher specificity and sensitivity, especially other molecular mechanism targets, such as immune infiltration, for improving the treatment of HNSCC. The aim of this study was to investigate the diagnostic value of SLA2 gene in HNSCC and its effect on tumor immune invasion. SLA2 is an adaptor protein whose main function is to inhibit the activation of activated T cell nuclear factor induced by T cell antigen receptor [21]. Relevant studies have reported that SLA2 expression mediates cell proliferation, colony formation and tumor formation by destroying receptors [3]. In conclusion, we can infer that SLA2 may be involved in tumor formation and development by further affecting cell proliferation through the cellular microenvironment.
In our study, we found that SLA2 was differentially expressed in various types of cancer and its corresponding normal tissues. A study indicates that SLA2 may be involved in immune response for promoting NSCLC progression. Besides, genome-scale analysis to identify prognostic markers in patients with early stage pancreatic ductal adenocarcinoma after pancreaticoduodenectomy, found that SLA2 is one of the prognostic markers [4]. The expression of SLA2 in HNSCC was further verified, and the results showed that patients with higher SLA2 expression had better prognosis. In addition, we found that SLA2 gene expression was associated with tumor stage, clinical grade, and age. In addition, univariate and multivariate Cox regression analysis showed that SLA2 expression was an independent prognostic factor for HNSCC. Therefore, SLA2 has potential diagnostic value in HNSCC.
Next, we analyzed the SLA2-related pathway in HNSCC to understand its carcinogenic mechanism. Based on GO and KEGG analyses, we observed that the functional network of SLA2 in HNSCC was related to the side of membrane, receptor complex, and other cellular component, where they were involved in biological processes, such as leukocyte cell–cell adhesion. These related genes also served as antigen binding, cytokine receptor activity, SH3 domain binding, and other molecular function cytokine receptor binding. Abplp is a yeast cortical actin binding protein that contains a SH3 domain similar to that found in signal transduction proteins that act at the membrane/cytoskeletal interface. Zero mutation of SLA1 and SLA2 leads to temperature sensitive growth defects. Sla1p contains three SH3 domains, which are essential for proper formation of cortical actin cytoskeleton [22]. KEGG pathway analysis indicated that these genes related to SLA2 were mainly enriched in signal pathways, such as hematopoietic cell lineage, CAMs, and other pathway. SLA2 is a recently discovered adaptor protein mainly expressed in hematopoietic cells. Studies shows that SLA2 may have function as a negative regulator of GPVI-mediated signaling by interacting with c-Cbl, similar to what was reported in T cells [23]. All these results suggested that SLA2 regulates immune effector process, and is involved in several related signaling pathways.
SLA2 is a adapter protein, which negatively regulates TCR signaling. It inhibits T-cell antigen–receptor-induced activation of nuclear factor of activated T-cells. It may act by linking signaling proteins, such as ZAP70 with CBL, leading to a CBL dependent degradation of signaling proteins. The study also shows that SLA2 is related to endosome membrane, which is involved in regulation of immune effector process [21]. In this study, we found that SLA2 expression levels were positively correlated with the infiltration levels of numerous immune cells, suggesting that SLA2 might promote the anti-tumor immune response in tumor tissues. Most malignancies increase levels of inhibitory ligands by impairing T cell function to avoid immune responses that lead to tumor growth. This is thought to be one of the potential mechanisms promoting tumor progression. Patients with higher levels of T-cell infiltration often have a better prognosis [24, 25]. In addition, SLA2 expression levels were significantly correlated with the expression levels of many immune molecules, which might recruit immune cell infiltration, thereby exerting an anti-tumor effect. Altogether, these results suggest that SLA2 may exert an anti-oncogenic function by triggering anti-tumor immune responses in HNSCC tissues.
Many reports support that immune cell infiltration in tumor tissues is related to tumor formation, growth and metastasis [26]. High concentration of immune promoting cells in tumor tissue can improve the prognosis of patients, while low concentration of immune promoting cells can lead to immune escape of cancer cells, resulting in poor prognosis [27]. When a tumor is strongly infiltrated by immune cells, the expression of SLA2 will increased, and strong infiltration by immune cells is associated with a more favourable prognosis. Our results are similar to the reported of Yang et al. [28].
To further evaluate the potential immune mechanisms of SLA2 in HNSCC, we second analyzed SLA2-related immune infiltration levels. The results demonstrate that the SLA2 expression level is strongly positive correlated with the infiltration level of many immune cells. After analyzing the correlation between SLA2 expression and immune cell marker genes, the results revealed that the function of SLA2 in regulating different immune infiltrating cells is different. SLA2 had a positive relationship with most of the immune activation genes. In addition, the efficacy of immunotherapy not only requires sufficient immune cell infiltration in the tumor microenvironment, but also depends on the full expression of immune checkpoints [29]. The results demonstrated that SLA2 positively co-expressed with these immune-checkpoints, indicating that SLA2 may be a potential immunotherapy target. Moreover, cisplatin and methotrexate had better antitumor effect for HNSCC.
A large number of studies have confirmed that tumor immune cell infiltration can affect the efficacy and prognosis of chemotherapy, radiotherapy or immunotherapy in cancer patients [30, 31]. Taken together, SLA2 expression level is closely related to immune cell infiltration, which can lead to good prognosis of tumor patients. Most importantly, SLA2 expression may affect the immune microenvironment and then indirectly affects the prognosis of HNSCC and serves as a predictor of ICB therapies and chemotherapeutics.However, the limitation of this study is that a large number of samples were needed to verify our results, and more clinical samples will be collected to enrich the data in the future. Moreover, the underlying immune mechanisms should be explored and SLA2 as biomarkers to predict the immune response rate in real-world HNSCC patients should be conducted.
In conclusion, our work demonstrated that when a tumor is strongly infiltrated by immune cells, the expression of SLA2 will increased, and strong infiltration by immune cells is associated with a more favourable prognosis. SLA2 may be a potential target for immunotherapy.
Data availability
The data in this study are available from the corresponding author upon request.
Abbreviations
- aDC:
-
Activated dendritic cell
- BP:
-
Biological process
- CAMs:
-
Cell adhesion molecules
- CBL:
-
Casitas B-lineage lymphoma proto-oncogene
- CC:
-
Cellular component
- CD274:
-
Cluster of differentiation 274
- CD4 + T cells:
-
Cluster of differentiation 4-positive T cells
- CD8 T cells:
-
Cluster of differentiation 8 T cells
- CD8 + T cells:
-
Cluster of differentiation 8-positive T cells
- CD28:
-
Cluster of differentiation 28 T cells
- CD48:
-
Cluster of differentiation 48 T cells
- CXCR4:
-
C–X–C motif chemokine receptor 4
- CTLA4:
-
Cytotoxic T lymphocyte-associated antigen-4
- DC:
-
Dendritic cell
- DSS:
-
Disease-specific survival
- GDSC:
-
The genomics of drug sensitivity in cancer
- GEPIA:
-
Gene expression profile interaction analysis
- GO:
-
Gene ontology
- GSEA:
-
Gene set enrichment analysis
- GSVA:
-
Gene set variation analysis
- GTeX:
-
The genotype-tissue expression
- HAVCR2:
-
Hepatitis A virus cellular receptor 2
- HNSCC:
-
Head and neck squamous cell carcinoma
- HR:
-
Hazard ratio
- ICB:
-
Immune checkpoint blockade
- IC50:
-
The samples’ half-maximal inhibitory concentration
- iDC:
-
Interdigitating dendritic cell
- IL6:
-
Interleukin-6
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- KIRC:
-
Kidney renal clear cell carcinoma
- KIRP:
-
Kidney renal papillary cell carcinoma
- KM:
-
Kaplan–Meier
- LAG3:
-
Lymphocyte-activation gene 3
- LCK:
-
Lymphocyte-specific protein tyrosine kinase
- MF:
-
Molecular function
- MHC genes:
-
Major histocompatibility complex gene
- mRNA:
-
Messenger ribonucleic acid
- NK CD56dim cells:
-
Natural killer CD56dim cells
- RNA-seq:
-
Ribonucleic acid sequencing
- SCC:
-
Squamous cell carcinoma
- SH3:
-
Src homology domain 3
- SIGLEC15:
-
Sialic acid binding Ig-like lectin 15
- SLA2:
-
Src-like adaptor 2 gene
- SsGSEA:
-
Single-sample gene set enrichment analysis
- STAD:
-
Stomach adenocarcinoma
- TCGA:
-
The cancer genome atlas
- TCR:
-
T-cell receptor
- Tcm:
-
Central memory T cells
- Tem:
-
Effective memory T cell
- TFH:
-
Follicular helper T cell
- Th1 cells:
-
T helper 1 cells
- Th2 cells:
-
T helper 2 cells
- TIGIT:
-
T cell immunoreceptor with Ig and ITIM domains
- TIICs:
-
Tumor-infiltrating immune cells
- TIMER:
-
Tumor Immune Estimation Resource
- TME:
-
Tumor microenvironment
- TPM:
-
Transcripts per million reads
- TReg:
-
Regulatory T cell
- OS:
-
Overall survival
- pDC:
-
Plasmacytoid dendritic cells
- PDCD1:
-
Programmed cell death 1
- PDCD1LG2:
-
Programmed cell death 1 ligand 2
- WHO:
-
World Health Organization
- ZAP70:
-
Zeta chain of T cell receptor-associated protein kinase 70
- 95% CI:
-
95% Confidence intervals
References
Sun X, Zhang L, Liu S (2021) The immune infiltration in hnscc and its clinical value: a comprehensive study based on the tcga and geo databases. Comput Math Methods Med 2021:1163250. https://doi.org/10.1155/2021/1163250
Meng Y, Huang T, Chen X, Lu Y (2021) A comprehensive analysis of the expression and regulation network of lymphocyte-specific protein tyrosine kinase in breast cancer. Transl Cancer Res. 10(3):1519–1536. https://doi.org/10.21037/tcr-21-328
Li J, Bi L, Shi Z, Sun Y, Lin Y, Shao H, Zhu Z (2016) Rna-seq analysis of non-small cell lung cancer in female never-smokers reveals candidate cancer-associated long non-coding rnas. Pathol Res Pract 212(6):549–554. https://doi.org/10.1016/j.prp.2016.03.006
Liao X, Huang K, Huang R, Liu X, Han C, Yu L, Yu T, Yang C, Wang X, Peng T (2017) Genome-scale analysis to identify prognostic markers in patients with early-stage pancreatic ductal adenocarcinoma after pancreaticoduodenectomy. Onco Targets Ther 10:4493–4506. https://doi.org/10.2147/ott.S142557
Cohen EEW, Bell RB, Bifulco CB, Burtness B, Gillison ML, Harrington KJ, Le QT, Lee NY, Leidner R, Lewis RL, Licitra L, Mehanna H, Mell LK, Raben A, Sikora AG, Uppaluri R, Whitworth F, Zandberg DP, Ferris RL (2019) The society for immunotherapy of cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (hnscc). J Immunother Cancer 7(1):184. https://doi.org/10.1186/s40425-019-0662-5
Wang G, Zhang M, Cheng M, Wang X, Li K, Chen J, Chen Z, Chen S, Chen J, Xiong G, Xu X, Wang C, Chen D (2021) Tumor microenvironment in head and neck squamous cell carcinoma: functions and regulatory mechanisms. Cancer Lett 507:55–69. https://doi.org/10.1016/j.canlet.2021.03.009
Jung AR, Jung CH, Noh JK, Lee YC, Eun YG (2020) Epithelial-mesenchymal transition gene signature is associated with prognosis and tumor microenvironment in head and neck squamous cell carcinoma. Sci Rep 10(1):3652. https://doi.org/10.1038/s41598-020-60707-x
Vivian J, Rao AA, Nothaft FA, Ketchum C, Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD, Musselman-Brown A, Schmidt H, Amstutz P, Craft B, Goldman M, Rosenbloom K, Cline M, O’Connor B, Hanna M, Birger C, Kent WJ, Patterson DA, Joseph AD, Zhu J, Zaranek S, Getz G, Haussler D, Paten B (2017) Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol 35(4):314–316. https://doi.org/10.1038/nbt.3772
Smyth GK, Michaud J, Scott HS (2005) Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21(9):2067–2075. https://doi.org/10.1093/bioinformatics/bti270
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z (2017) Gepia: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45(W1):W98-w102. https://doi.org/10.1093/nar/gkx247
Hänzelmann S, Castelo R, Guinney J (2013) Gsva: gene set variation analysis for microarray and rna-seq data. BMC Bioinformatics 14:7. https://doi.org/10.1186/1471-2105-14-7
Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman WH, Becker C, Pagès F, Speicher MR, Trajanoski Z, Galon J (2013) Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39(4):782–795. https://doi.org/10.1016/j.immuni.2013.10.003
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS (2017) Timer: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 77(21):e108–e110. https://doi.org/10.1158/0008-5472.Can-17-0307
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, Li Z, Traugh N, Bu X, Li B, Liu J, Freeman GJ, Brown MA, Wucherpfennig KW, Liu XS (2018) Signatures of t cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 24(10):1550–1558. https://doi.org/10.1038/s41591-018-0136-1
Lu X, Jiang L, Zhang L, Zhu Y, Hu W, Wang J, Ruan X, Xu Z, Meng X, Gao J, Su X, Yan F (2019) Immune signature-based subtypes of cervical squamous cell carcinoma tightly associated with human papillomavirus type 16 expression, molecular features, and clinical outcome. Neoplasia 21(6):591–601. https://doi.org/10.1016/j.neo.2019.04.003
Geeleher P, Cox NJ, Huang RS (2014) Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol 15(3):R47. https://doi.org/10.1186/gb-2014-15-3-r47
Jiang Q, Sun J, Chen H, Ding C, Tang Z, Ruan Y, Liu F, Sun Y (2021) Establishment of an immune cell infiltration score to help predict the prognosis and chemotherapy responsiveness of gastric cancer patients. Front Oncol 11:650673. https://doi.org/10.3389/fonc.2021.650673
Vasaikar SV, Straub P, Wang J, Zhang B (2018) Linkedomics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 46(D1):D956-d963. https://doi.org/10.1093/nar/gkx1090
Zhou LQ, Hu Y, Xiao HJ (2021) The prognostic significance of survivin expression in patients with hnscc: a systematic review and meta-analysis. BMC Cancer 21(1):424. https://doi.org/10.1186/s12885-021-08170-3
Oliva M, Spreafico A, Taberna M, Alemany L, Coburn B, Mesia R, Siu LL (2019) Immune biomarkers of response to immune-checkpoint inhibitors in head and neck squamous cell carcinoma. Ann Oncol 30(1):57–67. https://doi.org/10.1093/annonc/mdy507
Holland SJ, Liao XC, Mendenhall MK, Zhou X, Pardo J, Chu P, Spencer C, Fu A, Sheng N, Yu P, Pali E, Nagin A, Shen M, Yu S, Chan E, Wu X, Li C, Woisetschlager M, Aversa G, Kolbinger F, Bennett MK, Molineaux S, Luo Y, Payan DG, Mancebo HS, Wu J (2001) Functional cloning of src-like adapter protein-2 (slap-2), a novel inhibitor of antigen receptor signaling. J Exp Med 194(9):1263–1276. https://doi.org/10.1084/jem.194.9.1263
Holtzman DA, Yang S, Drubin DG (1993) Synthetic-lethal interactions identify two novel genes, sla1 and sla2, that control membrane cytoskeleton assembly in saccharomyces cerevisiae. J Cell Biol 122(3):635–644. https://doi.org/10.1083/jcb.122.3.635
Sugihara S, Katsutani S, Deckmyn H, Fujimura K, Kimura A (2010) Roles of src-like adaptor protein 2 (slap-2) in gpvi-mediated platelet activation slap-2 and gpvi signaling. Thromb Res 126(4):e276-285. https://doi.org/10.1016/j.thromres.2010.07.010
Oh DY, Kwek SS, Raju SS, Li T, McCarthy E, Chow E, Aran D, Ilano A, Pai CS, Rancan C, Allaire K, Burra A, Sun Y, Spitzer MH, Mangul S, Porten S, Meng MV, Friedlander TW, Ye CJ, Fong L (2020) Intratumoral cd4(+) t cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell 181(7):1612-1625.e1613. https://doi.org/10.1016/j.cell.2020.05.017
van der Leun AM, Thommen DS, Schumacher TN (2020) Cd8(+) t cell states in human cancer: Insights from single-cell analysis. Nat Rev Cancer 20(4):218–232. https://doi.org/10.1038/s41568-019-0235-4
Ringgaard L, Melander F, Eliasen R, Henriksen JR, Jølck RI, Engel TB, Bak M, Fliedner FP, Kristensen K, Elema DR, Kjaer A, Hansen AE, Andresen TL (2020) Tumor repolarization by an advanced liposomal drug delivery system provides a potent new approach for chemo-immunotherapy. Sci Adv. https://doi.org/10.1126/sciadv.aba5628
Chen H, Xie J, Jin P (2020) Assessment of hazard immune-related genes and tumor immune infiltrations in renal cell carcinoma. Am J Transl Res 12(11):7096–7113
Yang Z, Tian H, Bie F, Xu J, Zhou Z, Yang J, Li R, Peng Y, Bai G, Tian Y, Chen Y, Liu L, Fan T, Xiao C, Zheng Y, Zheng B, Wang J, Li C, Gao S, He J (2021) ERAP2 is associated with immune infiltration and predicts favorable prognosis in SqCLC. Front Immunol. https://doi.org/10.3389/fimmu.2021.788985
Chae YK, Arya A, Iams W, Cruz MR, Chandra S, Choi J, Giles F (2018) Current landscape and future of dual anti-ctla4 and pd-1/pd-l1 blockade immunotherapy in cancer; lessons learned from clinical trials with melanoma and non-small cell lung cancer (nsclc). J Immunother Cancer 6(1):39. https://doi.org/10.1186/s40425-018-0349-3
Zhang H, Liu H, Shen Z, Lin C, Wang X, Qin J, Qin X, Xu J, Sun Y (2018) Tumor-infiltrating neutrophils is prognostic and predictive for postoperative adjuvant chemotherapy benefit in patients with gastric cancer. Ann Surg 267(2):311–318. https://doi.org/10.1097/sla.0000000000002058
Lyu L, Yao J, Wang M, Zheng Y, Xu P, Wang S, Zhang D, Deng Y, Wu Y, Yang S, Lyu J, Guan F, Dai Z (2020) Overexpressed pseudogene hla-dpb2 promotes tumor immune infiltrates by regulating hla-dpb1 and indicates a better prognosis in breast cancer. Front Oncol 10:1245. https://doi.org/10.3389/fonc.2020.01245
Funding
This work was supported by State Administration of Traditional Chinese Medicine (Grant number: 201507006. Project name: Special Scientific Research project of TCM Industry of State Administration of Traditional Chinese Medicine), and Jiangxi Provincial Health and Family Planning Commission (Grant number:2018A375. Project name: Special Scientific Research project of TCM Industry of State Administration of Traditional Chinese Medicine).
Author information
Authors and Affiliations
Contributions
ZW and SH: conceived and designed the project. CY, ZZ, WW, JC, QX, CD and XH: performed the analyses of expression, survival prognosis, genetic alteration, gene enrichment, and immune infiltration. ZW, SH, CY and ZZ: wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that no confict of interest exists.
Ethical approval
Since all analyses in our study were mainly based on data from the TCGA database, ethical approval and patient written informed consent were not required.
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, Z., You, C., Zhu, Z. et al. SLA2 is a prognostic marker in HNSCC and correlates with immune cell infiltration in the tumor microenvironment. Eur Arch Otorhinolaryngol 281, 427–440 (2024). https://doi.org/10.1007/s00405-023-08213-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00405-023-08213-4