Abstract
Purpose
Human gene icb-1 recently has been reported to be part of a gene expression score predicting response to antiestrogen fulvestrant in breast cancer patients. In the present study, we examined to what extent icb-1 expression would affect the response of breast cancer cells to this antiestrogen in vitro and investigated underlying molecular mechanisms. Using open access mRNA data, we elucidated the significance of icb-1 expression for survival of breast cancer patients.
Methods
Icb-1 gene expression was knocked down by RNAi. Breast cancer cell growth after treatment with fulvestrant was assessed using the Cell Titer Blue assay. Gene expression was analyzed by Western blot analysis or RT-qPCR. Survival analyses were performed using bioinformatical online tools and data.
Results
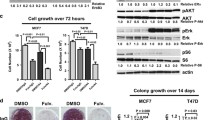
Knockdown of icb-1 in T-47D breast cancer cells significantly increased growth of this cell line and also elevated the growth-stimulatory effect of E2 (p < 0.001). After treatment with different concentrations of fulvestrant, icb-1 knockdown cells exhibited a significantly enhanced response to this drug (p < 0.01). On the molecular level, icb-1 knockdown led to elevated expression of ESR1 and its target gene TFF1 (pS2) and enhanced E2-triggered up-regulation of proliferation genes. Finally, bioinformatical meta-analysis of gene expression data of 3951 breast cancer patients revealed that high icb-1 expression increases their relapse-free survival (HR = 0.87, p < 0.05).
Conclusion
The presented data further support a tumor-suppressive role of icb-1 in breast cancer and suggest an inhibitory effect of this gene on fulvestrant action, which both are suggested to be mediated by suppression of cellular E2 response.






Similar content being viewed by others
Data availability
Data and materials are available from the corresponding author on reasonable request.
References
Treeck O, Strunck E, Vollmer G (1998) A novel basement membrane-induced gene identified in the human endometrial adenocarcinoma cell line HEC1B. FEBS Lett 425(3):426–430. https://doi.org/10.1016/s0014-5793(98)00278-6
Treeck O (2011) Silencing of the icb-1 gene inhibits the induction of differentiation-associated genes by vitamin D3 and all-trans retinoic acid in gynecological cancer cells. Int J Mol Med. https://doi.org/10.3892/ijmm.2011.663
Treeck O, Odani T, Itoh N et al (2002) Detection of increased icb-1 transcript levels in maturing HL-60 cells: a novel marker for granulocytic and monocytic in vitro differentiation. Leuk Res 26(8):765–769. https://doi.org/10.1016/s0145-2126(02)00005-x
Peirce MJ, Brook M, Morrice N et al (2010) Themis2/ICB1 is a signalling scaffold that selectively regulates macrophage toll-like receptor signalling and cytokine production. PLoS ONE 5(7):e11465. https://doi.org/10.1371/journal.pone.0011465
Muraoka S, Kume H, Adachi J et al (2013) In-depth membrane proteomic study of breast cancer tissues for the generation of a chromosome-based protein list. J Proteome Res 12(1):208–213. https://doi.org/10.1021/pr300824m
Treeck O, Kindzorra I, Pauser K et al (2005) Expression of icb-1 gene is interferon-gamma inducible in breast and ovarian cancer cell lines and affects the IFN gamma-response of SK-OV-3 ovarian cancer cells. Cytokine 32(3–4):137–142. https://doi.org/10.1016/j.cyto.2005.08.008
Bollmann J, Ortmann O, Treeck O (2008) Expression of differentiation-associated gene icb-1 is estrogen-responsive in ovarian and breast cancer cell lines. J Steroid Biochem Mol Biol 109(1–2):16–21. https://doi.org/10.1016/j.jsbmb.2007.12.007
Konwisorz A, Springwald A, Haselberger M et al (2010) Knockdown of ICB-1 gene enhanced estrogen responsiveness of ovarian and breast cancer cells. Endocr Relat Cancer 17(1):147–157. https://doi.org/10.1677/ERC-09-0095
Springwald A, Lattrich C, Seitz S et al (2009) Single nucleotide polymorphisms in human gene icb-1 and breast cancer susceptibility. Cancer Invest 27(6):669–672. https://doi.org/10.1080/07357900802620877
Treeck O, Schüler S, Häring J et al (2013) icb-1 Gene counteracts growth of ovarian cancer cell lines. Cancer Lett 335(2):441–446. https://doi.org/10.1016/j.canlet.2013.02.049
Treeck O, Belgutay D, Häring J et al (2012) Network analysis of icb-1 gene function in human breast cancer cells. J Cell Biochem 113(9):2979–2988. https://doi.org/10.1002/jcb.24175
Knudsen S, Jensen T, Hansen A et al (2014) Development and validation of a gene expression score that predicts response to fulvestrant in breast cancer patients. PLoS ONE 9(2):e87415. https://doi.org/10.1371/journal.pone.0087415
Girish V, Vijayalakshmi A (2004) Affordable image analysis using NIH Image/ImageJ. Indian J Cancer 41(1):47
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3(6):1101–1108. https://doi.org/10.1038/nprot.2008.73
Tang Z, Li C, Kang B et al (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45(W1):W98–W102. https://doi.org/10.1093/nar/gkx247
Györffy B, Lanczky A, Eklund AC et al (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123(3):725–731. https://doi.org/10.1007/s10549-009-0674-9
Perou CM, Sørlie T, Eisen MB et al (2000) Molecular portraits of human breast tumors. Nature 406(6797):747–752. https://doi.org/10.1038/35021093
Sorlie T, Perou CM, Tibshirani R et al (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci 98(19):10869–10874. https://doi.org/10.1073/pnas.191367098
Ikeda K, Horie-Inoue K, Inoue S (2015) Identification of estrogen-responsive genes based on the DNA binding properties of estrogen receptors using high-throughput sequencing technology. Acta Pharmacol Sin 36(1):24–31. https://doi.org/10.1038/aps.2014.123
Blanchard JM (2000) Cyclin A2 transcriptional regulation: modulation of cell cycle control at the G1/S transition by peripheral cues. Biochem Pharmacol 60(8):1179–1184. https://doi.org/10.1016/s0006-2952(00)00384-1
Sales Gil R, Vagnarelli P (2018) Ki-67: more hidden behind a “classic proliferation marker.” Trends Biochem Sci 43(10):747–748. https://doi.org/10.1016/j.tibs.2018.08.004
Acknowledgements
We would like to thank Mrs. Bettina Federhofer for expert technical assistance.
Funding
None.
Author information
Authors and Affiliations
Contributions
MR: data collection and management, and data analysis. MS, SS: data analysis and manuscript editing. OO: providing essential project support and manuscript editing. OT: protocol/project development, data collection and management, data analysis, and manuscript writing/editing.
Corresponding author
Ethics declarations
Conflict of interest
All authors state that they do not have any conflicts of interest with regard to this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Riedmeier, M., Skrzypczak, M., Schüler-Toprak, S. et al. Icb-1 expression inhibits growth and fulvestrant response of breast cancer cells and affects survival of breast cancer patients. Arch Gynecol Obstet 304, 203–213 (2021). https://doi.org/10.1007/s00404-020-05902-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00404-020-05902-x