Abstract
The late cornified envelope (LCE) gene cluster is located on chromosome 1q21, including LCE1–LCE6. Several single nucleotide polymorphisms (SNPs) in the LCE cluster were associated with susceptibility to psoriasis in Chinese population. However, there is no report on the relationship in ethnic minority areas in China. This study aimed to investigate the association between the gene polymorphisms of LCE1B, LCE1C, LCE3A, LCE3D and psoriasis vulgaris among Mongolians from Inner Mongolia. Totally, 305 Mongolians with psoriasis vulgaris (PsV) and 383 healthy controls were enrolled in the study from 2006 to 2015. 7 SNPs including rs6701216, rs4112788, rs12023196, rs512208, rs4845454, rs4085613 and rs1886734, were selected for genotyping with ligase detection reaction (LDR). Statistical analysis was performed for comparisons of allele frequencies and genotype frequencies between the patient group and the control group. In this study, excluding rs4085613 and rs1886734, differences were detected in the allele frequencies of other 5 SNPs between the patients and controls. Genotype analysis showed that under the recessive inheritance model, the genotype frequencies of rs4845454, rs4112788 differed between the patients and controls (all p < 0.00 5).Under the dominant and the recessive model, the genotype frequencies of rs6701216, rs12023196 and rs512208 significantly differed between the patients and controls. The LD analysis showed that strong LD existed between rs6701216 and rs12023196, rs4845454 and rs4085613, rs4845454 and rs1886734, and rs4085613 and rs1886734. The SNPs rs6701216, rs4112788, rs12023196, rs512208 and rs4845454 in the LCE gene were associated with psoriasis vulgaris among Mongolians from Inner Mongolia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Psoriasis is a common, recurrent, chronic, inflammatory skin disease with a prevalence of 0.47% in Chinese [12, 16]. It is characterized by high epidermis proliferation, abnormal keratinocytes differentiation and inflammatory reaction [16]. The pathogenesis of this complicated disease remains unclear. Recent studies suggest that genetic factors have relatively important influence on the pathogenesis of psoriasis. Since the nineties of the last century, with the development of molecular biology technology, the research on psoriasis susceptibility genes has shown promising results. Notably, completion of the human genome project (HGP) has resulted in landmark progress in the study of genetic diseases such as psoriasis. But psoriasis is a complex genetic disease, which is affected by genetic factors, geographical, ethnic and environmental stimulation. Therefore, to explore the pathogenesis and find the susceptible genes of psoriasis, it is also necessary to investigate different populations worldwide. Mongolians are an important group in north China. As compared to other nationalities, Mongolians have unique genetic background, living environment and living habits. Therefore, study on susceptibility genes of psoriasis in Mongolians will also provide valuable data on the genetics of psoriasis and more theoretical basis for the diagnosis and treatment of psoriasis at the gene level in the future.
Since 2008, a large number of susceptible genes of psoriasis were found [2, 6]. The LCE gene is located in the epidermal differentiation complex region of chromosome 1q21.3 [4], which can be further divided into six subgroups [17]. Among them, LCE3 gene cluster contains LCE3A, LCE3B, LCE3C, LCE3D and LCE3E, which have different structure and function [5]. The association between LCE and psoriasis was confirmed in different ethnic groups [1, 8, 15]. In 2008, new susceptibility loci for psoriasis-LCE1C (rs6701216) was found through GWAS in Europeans and Americans [9]. In 2009, LCE3D (rs4112788, rs4085613) and LCE3A (rs4845454, rs1886734) [8] was reported to be associated with psoriasis in Han and Uygur. In addition, this research also confirmed that LCE1B (rs12023196) may be associated with susceptibility of psoriasis in Chinese Han population. Huayang Tang [9] et al. confirmed that rs512208 in LCE3D was significantly associated with psoriasis in Chinese. In our study, seven single nucleotide polymorphisms (SNPs) in the LCE region, which are mentioned above, were chosen to explore their relationship with the Mongolian psoriasis vulgaris in Inner Mongolia of China.
Materials and methods
Materials
Subjects and controls
From January 2006 to December 2015, a total of 305 psoriasis vulgaris and 383 healthy controls were enrolled into the study. All patients and controls were Mongolians. The patients were recruited from the Dermatology department of the Affiliated Hospital of Inner Mongolia Medical University, in accordance with the diagnostic criteria of psoriasis vulgaris after being diagnosed by four clinical dermatologists through clinical or pathological methods. All controls were healthy people who had undergone physical examination in the Affiliated Hospital of Inner Mongolia Medical University. The inclusion criteria for controls were: (1) no autoimmune and systemic diseases, (2) no history of psoriasis, (3) no family history of psoriasis in one, two, three degree relatives. All controls were gender-matched with the psoriasis patients. More than three generations of the subjects and controls in our study had been living in Inner Mongolia and were not related to each other.
All patients and controls accepted a rigorous questionnaire survey and follow-up with informed consent. The study was approved by the ethics committee of the Affiliated Hospital of Inner Mongolia Medical University (Ethical approval number: 2007-001). The investigations were conducted according to the Declaration of Helsinki principles.
Selection of SNPs
Based on previous reports, 7 SNPs including rs6701216, rs4112788, rs12023196, rs512208, rs4845454, rs4085613 and rs1886734 in the LCE region were selected in this study3,5,7,8,13, 14, 15.
Methods
Genomic DNA extraction
Two milliliters of venous blood samples were collected from each subject after signing informed consent. Edtap dipotassium ethylene diamine tetraacetate (EDTAK2) was added into the blood samples as anticoagulant. All samples were stored at − 80 °C. AxyPrep-96 Whole blood genomic DNA kit from AXYGEN (33210 Central Avenue Union City, California, 94587, USA) was used to extract genomic DNA from patients and controls. The absorbance at 260 and 280 nm were detected by spectrophotometer. The concentration of DNA was calculated. The integrity of DNA was detected by 0.8% agarose gel electrophoresis.
Synthesis of primers and probes
Primer 3 online (Version 0.4.0) (http://frodo.wi.mit.edu/) and oligo (Version 6.31) (Molecular biology insights Inc, USA) softwares were used to design LDR probes and specific primers. Two primers were designed for each SNP locus. We tried our best to ensure that all SNP loci were in the same T value. Polymerase chain reaction (PCR) primer sequences are shown in Table 1. The upstream and downstream probes of the ligase detection reaction (LDR) were designed according to the design principle of LDR probe [3, 7]. The 5′end of the upstream probe was modified by phosphorylation. The LDR probe sequence is shown in the animation (Online Resource).
Multiplex PCR
PCR reaction system was 20 µl, which consisted of the following components: 1 µl templates, 2 µl buffer, 0.6 µl Mg2+, 2 µl dNTP, 0.2 µl TaqDNA polymerases, 2 µl primer mix and 12.2 µl H2O. The cycling conditions included initial denaturation at 95 °C for 15 min, 35 cycles of three steps (denaturation at 94 °C for 30 s, annealing at 56 °C for 1 min and extension at 65 °C for 30 s), followed by a final extension at 65 °C for 10 min.
Multiplex LDR
The ligation reaction was performed in a final volume of 10 µl containing 1 µl buffer, 1 µl probe mixture, 0.05 µl Taq DNA ligase, 4 µl PCR product and 3.95 µl H2O. Probe mixture concentration was 2 pmol/µl. The cycling conditions included denaturation at 95 °C for 2 min, 40 cycles of two steps (annealing at 94 °C for 15 s and extension at 50 °C for 25 s).
Sequencing and genotyping
The reaction products were sequenced and analyzed using PRISM 3730 sequencer. Finally, data were analyzed using GeneMapper software. The sequencing was completed by the Shanghai Yihe application of Biotechnology Co. Ltd.
Statistical analysis
Statistical analyses were performed using PLINK 1.07 software (http://pngu.mgh.harvard.edu/purcell/plink/) [3]. All allele and genotype frequencies of 7 SNPs in the case and control groups were calculated, and the Hardy–Weinberg genetic equilibrium analysis was performed. Chi-square test (χ2) was used to compare the allele and genotype frequencies. Differences in allele frequencies were quantified by OR and 95% CI using SPSS for Windows software (version 17.0; SPSS, Chicago, IL, USA). Linkage disequilibrium between the 7 SNPs was analyzed, and the values of R2 and D were calculated. All P values were two-sided, and p < 0.05 was considered to be statistically significant, p < 0.007 (0.05/7) was considered to achieve the Bonferroni multiple test correction level.
Results
Sample and SNP quality control
A total of 305 psoriasis vulgaris (mean age 40.20 ± 15.67, 171 males and 134 females) patients and 383 healthy controls (mean age 28.71 ± 12.36, 181 males and 202 females) were enrolled into the study. There was no significant difference in gender and age distribution between the patient and control groups (p > 0.05). The distribution of the 7 SNPs was in accordance with the Hardy–Weinberg equilibrium in both groups (p > 0.1).
Allele frequency comparison (Table 2)
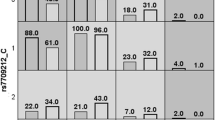
The linkage disequilibrium (LD) analysis between every 2 SNPs showed strong LD between rs6701216 and rs12023196 (R2 = 0.720, D = 0.903), rs4845454 and rs4085613 (R2 = 0.681, D = 0.857), rs4845454 and rs1886734 (R2 = 0.705, D = 0.849), and rs4085613 and rs1886734 (R2 = 0.680, D = 0.847) (Fig. 1).
Genotype frequency comparison
Genotype analysis showed that under the recessive inheritance model, the genotype frequencies of rs4845454, rs4112788 differed between the patients and controls, all of which can reach the level of Bonferroni correction(all p < 0.00 7).Under the dominant and the recessive model, the genotype frequencies of rs6701216, rs12023196 and rs512208 significantly differed between the patients and controls. (Table 3). In addition, we also used the logical regression method to analyze the additive models of the 7 SNPs. The results showed that under the additive model, the P values of rs6701216, rs12023196, rs512208 and rs4112788 were less than 0.05, all of which can reach the level of Bonferroni correction(all p < 0.00 7), indicating that these 4 SNPs were significantly correlated with psoriasis (Table 4).
Discussion
In this study, we replicated the previously reported association of 7 SNPs with Mongolian psoriasis in Inner Mongolia. Ying Liu et al. confirmed that the expression frequency of LCE1C (rs6701216) in Caucasians with psoriasis was significantly higher than that in the control group, and the allele T was associated with the risk of psoriasis [9], which is consistent with our results. 4 SNP loci including rs4112788, rs4845454, rs1886734, and rs4085613 were found to be significantly associated with psoriasis in Chinese Han and Uygur by Zhang Xuejun [8]. We further detected these 4 susceptibility loci in 305 Mongolians with psoriasis vulgaris in Inner Mongolia of China. However, differences were detected in the allele frequencies of only 2 SNPs (rs4112788, rs4845454) between the patients and controls. The other 2 SNPs (rs4085613、rs1886734) of them were confirmed having no significant difference between patients and controls in our study. Besides, Zhang Xuejun [8] showed that the correlation between SNP rs6701216 and Chinese psoriasis patients was very weak (p = 0.02), which is very different from our study. We analyzed that the results may be affected by race and environmental factors, which need to further expand the sample for validation. We used the ligase detection reaction for rs12023196 genotyping in 305 Mongolians with psoriasis vulgaris and 383 normal controls, which showed that the allele frequencies in the case and control groups had significant differences (p < 0.001, OR = 1.723, 95% CI = 1.384–2.144). Besides, the allele T of rs12023196 was associated with the risk of psoriasis. Prior to this, Zhang Xuejun et al. had also arrived at the same conclusion in a genetic study of 1103 Chinese Han patients with psoriasis. In this study, we confirmed that LCE3D (rs512208) was significantly associated with Mongolian psoriasis in Inner Mongolia, which was consistent with the findings of Huayang Tang [9]. The results of this study showed that the 5 SNPs in LCE gene were related to Mongolian psoriasis in Inner Mongolia. The linkage disequilibrium (LD) analysis showed strong LD between rs6701216 and rs12023196, rs4845454 and rs4085613, rs4845454 and rs1886734, and rs4085613 and rs1886734. Genotype analysis showed that the allele C of rs4845454, allele A of rs4085613 and allele T of rs1886734 may be recessive. Besides, the analysis of the additive models of these 7 loci can further confirm that rs6701216, rs12023196, rs512208 and rs4112788 were significantly correlated with psoriasis vulgaris among Mongolians from Inner Mongolia.
In conclusion, this study confirmed the association of LCE gene polymorphism with Mongolian psoriasis in Inner Mongolia through genetic methods, and further proved that the LCE gene may play an important role in the pathogenesis of psoriasis, providing new insights into the genetics of psoriasis. But there are some limitations in this research. First, this study mainly focused on the association analysis of single gene polymorphisms, and did not explore the complicated interaction between LCE gene polymorphisms and other genes or clarify whether the association between genetic variation and disease susceptibility was affected by other genes. Second, how LCE polymorphisms affect its gene function and immune system function needs more in-depth study in the future.
References
Bergboer JG, Tjabringa GS, Kamsteeg M, van Vlijmen-Willems IM, Rodijk-Olthuis D, Jansen PA, Thuret JY, Narita M, Ishida-Yamamoto A, Zeeuwen PL, Schalkwijk J (2011) Psoriasis risk genes of the late cornified envelope-3 group are distinctly expressed compared with genes of other LCE groups. Am J Pathol 178:1470–1477. https://doi.org/10.1016/j.ajpath.2010.12.017
Chandran V (2013) The genetics of psoriasis and psoriatic arthritis. Clin Rev Allergy Immunol 44:149–156. https://doi.org/10.1007/s12016-012-8303-5
Duffin KC, Chandran V, Gladman DD, Krueger GG, Elder JT, Rahman P (2008) Genetics of psoriasis and psoriatic arthritis: update and future direction. J Rheumatol 35:1449–1453
Jackson B, Tilli CM, Hardman MJ, Avilion AA, MacLeod MC, Ashcroft GS, Byrne C (2005) Late cornified envelope family in differentiating epithelia-response to calcium and ultraviolet irradiation. J Investig Dermatol 124:1062–1070. https://doi.org/10.1111/j.0022-202X.2005.23699.x
Liu Y, Helms C, Liao W, Zaba LC, Duan S, Gardner J, Wise C, Miner A, Malloy MJ, Pullinger CR, Kane JP, Saccone S, Worthington J, Bruce I, Kwok PY, Menter A, Krueger J, Barton A, Saccone NL, Bowcock AM (2008) A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet 4:e1000041. https://doi.org/10.1371/journal.pgen.1000041
Mischke D, Korge BP, Marenholz I, Volz A, Ziegler A (1996) Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (epidermal differentiation complex) on human chromosome 1q21. J Investig Dermatol 106:989–992
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1086/519795
Shen C, Gao J, Yin X, Sheng Y, Sun L, Cui Y, Zhang X (2015) Association of the late cornified envelope-3 genes with psoriasis and psoriatic arthritis: a systematic review. J Genet Genom Yi chuan xue bao 42:49–56. https://doi.org/10.1016/j.jgg.2015.01.001
Tang H, Jin X, Li Y, Jiang H, Tang X, Yang X, Cheng H, Qiu Y, Chen G, Mei J, Zhou F, Wu R, Zuo X, Zhang Y, Zheng X, Cai Q, Yin X, Quan C, Shao H, Cui Y, Tian F, Zhao X, Liu H, Xiao F, Xu F, Han J, Shi D, Zhang A, Zhou C, Li Q, Fan X, Lin L, Tian H, Wang Z, Fu H, Wang F, Yang B, Huang S, Liang B, Xie X, Ren Y, Gu Q, Wen G, Sun Y, Wu X, Dang L, Xia M, Shan J, Li T, Yang L, Zhang X, Li Y, He C, Xu A, Wei L, Zhao X, Gao X, Xu J, Zhang F, Zhang J, Li Y, Sun L, Liu J, Chen R, Yang S, Wang J, Zhang X (2014) A large-scale screen for coding variants predisposing to psoriasis. Nat Genet 46:45–50. https://doi.org/10.1038/ng.2827
Tang X, Li M, Zhu C (2009) Association of polymorphism of LCE3D gene with psoriasis vulgaris in Chinese Han population. J Anhui Med College 44:337–340. https://doi.org/10.3969/j.issn.1000-1392.2009.03.016
Thomas G, Sinville R, Sutton S, Farquar H, Hammer RP, Soper SA, Cheng YW, Barany F (2004) Capillary and microelectrophoretic separations of ligase detection reaction products produced from low-abundant point mutations in genomic DNA. Electrophoresis 25:1668–1677
Xiao L, Wang T, Shen Y (2010) Epidemiological survey of psoriasis in six provinces of China. Chin J Dermatovenereol 24:598–601
Zhang P, Zhu Y, Chen L (2011) Expression and significance of 1B and LCE3D in the skin lesions of psoriasis with the late stage of the stratum corneum envelope protein (LCE). J Clin Dept Dermatol 40:666–668. https://doi.org/10.3969/j.issn.1000-4963.2011.11.007
Zhang X, Zhang C, Ren W (2009) Association of LCE1B gene polymorphism with psoriasis in Chinese Han population. J Anhui Med College 44:311–314. https://doi.org/10.3969/j.issn.1000-1492.2009.03.008
Zhang XJ, Huang W, Yang S, Sun LD, Zhang FY, Zhu QX, Zhang FR, Zhang C, Du WH, Pu XM, Li H, Xiao FL, Wang ZX, Cui Y, Hao F, Zheng J, Yang XQ, Cheng H, He CD, Liu XM, Xu LM, Zheng HF, Zhang SM, Zhang JZ, Wang HY, Cheng YL, Ji BH, Fang QY, Li YZ, Zhou FS, Han JW, Quan C, Chen B, Liu JL, Lin D, Fan L, Zhang AP, Liu SX, Yang CJ, Wang PG, Zhou WM, Lin GS, Wu WD, Fan X, Gao M, Yang BQ, Lu WS, Zhang Z, Zhu KJ, Shen SK, Li M, Zhang XY, Cao TT, Ren W, Zhang X, He J, Tang XF, Lu S, Yang JQ, Zhang L, Wang DN, Yuan F, Yin XY, Huang HJ, Wang HF, Lin XY, Liu JJ (2009) Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat Genet 41:205–210. https://doi.org/10.1038/ng.310
Zhao B (2010) Chinese Journal of clinical dermatology. Jiangsu Science and Technology Press, Nanjing
Zhao XP, Elder JT (1997) Positional cloning of novel skin-specific genes from the human epidermal differentiation complex. Genomics 45:250–258. https://doi.org/10.1006/geno.1997.4952
Acknowledgements
This study was supported by Major Project of Affiliated Hospital of Inner Mongolia Medical University (No. NYFYZD2012007).
Funding
This study was supported by Major Project of Affiliated Hospital of Inner Mongolia Medical University (No. NYFYZD2012007).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants
The study was approved by the ethics committee of the Affiliated Hospital of Inner Mongolia Medical University (Ethical approval number: 2007-001). The investigations were conducted according to the Declaration of Helsinki principles.
Informed consent
All patients and controls accepted a rigorous questionnaire survey and follow-up with informed consent.
Additional information
Li Sun, Yuting Cao and Nagonbilig He contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Sun, L., Cao, Y., He, N. et al. Association between LCE gene polymorphisms and psoriasis vulgaris among Mongolians from Inner Mongolia. Arch Dermatol Res 310, 321–327 (2018). https://doi.org/10.1007/s00403-018-1813-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00403-018-1813-0