Abstract
We investigated the effects of substrate (cellulose or starch) and different clay contents on the production of microbial extracellular polymeric substances (EPS) and concomitant development of stable soil aggregates. Soils were incubated with different amounts of montmorillonite (+ 0.1%, + 1%, + 10%) both with and without two substrates of contrasting quality (starch and cellulose). Microbial respiration (CO2), biomass carbon (C), EPS-protein, and EPS-polysaccharide were determined over the experimental period. The diversity and compositional shifts of microbial communities (bacteria/archaea) were analysed by sequencing 16S rRNA gene fragments amplified from soil DNA. Soil aggregate size distribution was determined and geometric mean diameter calculated for aggregate formation. Aggregate stabilities were compared among 1–2-mm size fraction. Starch amendment supported a faster increase than cellulose in both respiration and microbial biomass. Microbial community structure and composition differed depending on the C substrate added. However, clay addition had a more pronounced effect on alpha diversity compared to the addition of starch or cellulose. Substrate addition resulted in an increased EPS concentration only if combined with clay addition. At high clay addition, starch resulted in higher EPS concentrations than cellulose. Where additional substrate was not provided, EPS-protein was only weakly correlated with aggregate formation and stability. The relationship became stronger with addition of substrate. Labile organic C thus clearly plays a role in aggregate formation, but increasing clay content was found to enhance aggregate stability and additionally resulted in the development of distinct microbial communities and increased EPS production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil health and functioning are linked to the stability of soil structure (Doetterl et al. 2016), which in turn is the key to organic matter (OM) stabilisation (Lehmann and Kleber 2015). The formation, stabilisation, and destruction of soil aggregates occur through interactions between organic and inorganic soil constituents and other environmental factors, e.g. climate and vegetation (Kiem and Kandeler 1997; Six et al. 2004). By the conceptual formulation of the aggregate hierarchy model, Oades (1984) proposed that microorganisms decompose OM within macroaggregates, which subsequently becomes encapsulated with minerals and microbial residues to form microaggregates within macroaggregates. Hence, aggregate turnover (the rate of aggregate formation and destruction) is anticipated to be strongly influenced by microbial activity. However, the knowledge on the interactions between microbial processes, soil minerals, and the availability of OM for decomposition is still very limited.
While a role of microorganisms in soil aggregation has long been recognised (Chenu and Cosentino 2011; Martin and Waksman 1940), the relative contribution of microbial parameters such as microbial biomass, activity, microbial community composition, and the secretion of biological bonding agents, such as extracellular polymeric substances (EPS), is not well understood. Fungal hyphae physically enmesh and connect microaggregates together, forming larger macroaggregates, but they also stabilise the material within microaggregates (Six et al. 2004), suggesting fungal exudates also have a role (e.g. Wright and Upadhyaya (1998)). De Gryze et al. (2005) presented mixed reports, indicating that aggregate formation and stability could sometimes correlate with microbial activity and biomass, and sometimes not. Helfrich et al. (2015) found that the amount of fungal phospholipid fatty acids (PLFA) was related to the abundance of water-stable macroaggregates. The authors suggested that not only fungal biomass but also fungal activity was of importance in the formation of macroaggregates. On the contrary, Guggenberger et al. (1999) observed that the content of macroaggregates did not decline with a decrease in either bacterial or fungal biomass and concluded that while living organisms may create macroaggregates, other factors induced by microorganisms were involved in their stabilisation. These studies suggest that the response of aggregate formation to one microbial parameter, e.g. biomass could be a function of conjoint interactions with abiotic parameter(s). For instance, De Gryze et al. (2005) suggested aggregate stabilisation might be more texture-dependent than aggregate formation. The authors observed that the amount of water-stable aggregates (> 2 mm) in natural ecosystems decreased in the order silty clay loam > silt loam > sandy loam and no significant effects of soil texture on macroaggregate formation after addition of wheat residue. Kiem and Kandeler (1997) found that the increase in aggregate stability induced by the microbial biomass was greatest in sandy soils (< 15% clay) and least in clayey soils (> 35% clay). Additionally, clay minerals may exert stress indirectly on microbes, by affecting extracellular microbial enzymes through adsorption (Olagoke et al. 2019, 2020; Quiquampoix et al. 2002). It has been shown that bacterial exposure to mineral surfaces (kaolinite and goethite) can cause physical stress and mortality (Ma et al. 2017). As a survival strategy, this may result in high microbial EPS production in soils with higher clay content that would in turn be significant for soil aggregate formation and stability.
The production of binding substances by microorganisms has been proposed as one key factor influencing aggregate stability (Kiem and Kandeler 1997). According to Costa et al. (2018), the production of EPS is being acknowledged as a binding agent and one important driver of soil aggregation. EPS facilitate the initial attachment of cells to various substrates and protect against environmental stresses and dehydration (Wingender et al. 1999). As growth and colony formation take place, more EPS is produced, creating adhesives that glue adjacent clay particles together to form aggregates (Wright and Upadhyaya 1998). Depending on the decomposability of OM, microbes might differ in their EPS production as consequence of increasing activity and variation in carbon source (Sheng et al. 2006). Accordingly, Vogel et al. (2014) proposed that a heterogeneous mixture of EPS (e.g. differing in amount and/or quality) released by diverse microbial communities might glue soil particles together to different extents and possibly create a basis for differing influence on soil aggregation. Indeed, Redmile-Gordon et al. (2020) found that EPS concentration and soil structural stability were greater in soils under diverse perennial plant cover than under arable monoculture — with EPS-protein showing a closer relationship than EPS-polysaccharide to the observed aggregate stability. However, the aforementioned study did not investigate the effects of clay content on this relationship nor track short-term changes in aggregate formation and dispersal. Therefore, while EPS-proteins with their functional groups (e.g. amino and carboxylic groups) interacting with soil mineral surfaces are anticipated to be more influential for aggregate stability than polysaccharides — little is known about how substrate quality input and clay content interact to influence the resulting parameters.
This study aimed to evaluate the influences of (i) substrate quality, (ii) soil texture, and (iii) the microbial properties (that is, community composition, biomass, and their EPS production) on soil aggregate formation and stability. Our overarching goal was to evaluate which among these factors define the temporal patterns of aggregate turnover. To understand the role of microorganisms in aggregate turnover, we have considered their abundance, composition, and activity. We evaluated the effect of soil texture (by varying soil clay content) and contrasting carbon (C) substrate quality (starch vs. cellulose) on microbial community composition and activities and measured the effects on soil aggregate formation and stability. We hypothesised that (1) molecular differences in the added organic substrate will largely drive the composition of the microbial community — but interacting with clay content. (2) Substrate addition will stimulate microbial production of EPS, and the more labile substrate (starch) will result in a faster microbial response compared to the more chemically recalcitrant substrate (cellulose). (3) Microbial EPS production will depend on the clay content, with higher clay contents triggering greater EPS production. (4) Even though clay itself will have a positive influence on aggregation, any clay-driven increases in EPS production will be reflected by accordingly increased soil aggregate formation and stability.
Materials and methods
Soil material and incubation experiment
A sandy soil from the depositional area of an erosion field experiment (conventional tillage) from Müncheberg (ZALF-Leibniz Centre for Agricultural Landscape Research, Germany) was sampled and sieved to < 2 mm, with large organic residues and roots being removed. The soil textural component consisted of sand 86.4%, silt 9.2%, and clay 4.4%. Detailed characteristics of the soil are described by Olagoke et al. (2019). The soils were pre-incubated twice before the main incubation. For the first pre-incubation, soil was incubated at 20 °C for 14 days to facilitate break-down of large macroaggregates by exhaustion of labile OM. The soil was then air-dried at 38 °C and aggregates were disrupted by hand. The particulate OM which becomes available due to aggregate disruption was removed by electrostatic attraction (Kaiser et al. 2009) to reduce the amount of labile C. The resulting disaggregated soil was modified with different amounts of montmorillonite (montmorillonite–CERATOSIL® WGD fein), to simulate a gradient in clay content: increasing in a logarithmic scale from + 0.1 and + 1 to + 10%. As the clay mineral addition increased the pH of the soil which was originally 5.9, Ca(OH)2 was used to achieve a uniform pH of 7 for all treatments. After remoistening, the four different treatments (original soil without clay addition, + 0.1%, + 1%, + 10% clay addition) were pre-incubated again for ca. 24 weeks at 20 °C, allowing the microorganisms adapt to the new conditions. Then, the soil material was air-dried and macroaggregates were destroyed by sieving < 250 µm.
For the main incubation experiment, 200 g of each modified soil were placed into 850 ml incubation jars. Starch (readily available C; 1 mg C g−1 soil), cellulose (relatively recalcitrant; 1 mg C g−1 soil), or no substrate was added in factorial combination with the four contrasting clay contents to test the effect of OM decomposability on aggregation. A conservative addition of ammonium nitrate (10 µg N g−1 soil) was included to prevent excessive N limitation while ensuring a relatively high CN ratio, which was previously shown to promote EPS production (Redmile-Gordon et al. 2015). Water was added to reach 50% of the maximum water holding capacity and the soils were gently mixed. Water was added as needed to maintain this water content. Soils were incubated at 20 °C in the dark for 80 days and were destructively sampled at 0, 3, 10, 20, 40, and 80 days (Fig. 1). All treatments were run in four independent replicates per treatment and time step.
Determination of microbial parameters
Microbial activity and biomass C
The microbial activity was measured as CO2 respiration by titration over the incubation time. To provide feedback on C mineralisation, 25 ml vials containing aqueous NaOH were placed in the middle of the incubation jars to trap the CO2. In addition, eight incubation jars containing only NaOH were used as blanks to correct for the CO2 trapped from the air inside the vessels. The evolved CO2 was captured in 0.05 N NaOH for blanks as well as controls (no substrate addition) and in 0.1 to 0.4 N NaOH for treatments with substrate addition depending on the expected microbial activity based on the previous measurement. The NaOH solution was exchanged after approximately 6 and 18 h at the beginning of the incubation, and subsequently after longer time steps adapted depending on the former microbial activity until day 79. At the time of replacement, BaCl2 was used for precipitation with a subsequent titration against HCl. Phenolphthalein was used as the indicator in the titration reaction, whereby the titration factor was determined in triplicate. After changing NaOH solution, all incubation jars were kept open for 10 min to replenish O2.
Microbial biomass C was measured by fumigation-extraction (Vance et al. 1987; Wu et al. 1990) by determining C extracted from 5 g of fumigated vs. non-fumigated soil, for all treatments and sampling times (n = 288). The fumigated aliquot was incubated in a desiccator for 24 h at 25 °C. Then, the chloroform was completely removed by evacuating the desiccator several times. Thereafter, 20 ml of 0.05 M K2SO4 was added to both fumigated and non-fumigated aliquots and shaken for 60 min using an end-over-end shaker at 35 rpm. The extracts were then filtered through a Whatman filter paper (no. 42, Sigma-Aldrich, Germany). Organic C (OC) and N content in both extracts were measured using a Vario TOC cube (Elementar, Germany). The microbial biomass C was calculated as the difference in extractable OC contents between fumigated (OCf) and non-fumigated samples (OCnf) considering 0.45 as a factor of microbial biomass extraction efficiency (kEC) (Jenkinson et al. 2004) following Eq. 1.
EPS extraction and analyses
The EPS was extracted from all soils (n = 288) following the protocol by Frølund et al. (1996) adapted for soils by Redmile-Gordon et al. (2014). Briefly, 2.5 g of fresh soil was placed in centrifuge tubes, capped to minimise the evaporation, and kept at 4 °C throughout the extraction procedure. After the removal of soluble constituents using CaCl2 solution, EPS was extracted from the residual soil by shaking with 2.5 g cation exchange resin (Sigma-Aldrich /DOWEX, Saint Louis, USA, PN 91,973) in an extraction buffer. Extracts were immediately stored at − 20 °C until analyses. The total saccharide content from extracted EPS (EPS-polysaccharide) was quantified after DuBois et al. (1956) using D( +)-Glucose as a standard. The protein content was determined using a modified microplate technique (adapted from Lowry et al. (1951) by Redmile-Gordon et al. (2013)) using bovine serum albumin as a standard. This method was chosen because colorimetric analysis of protein in soil extracts can be affected by chemical interferences, especially polyphenolics (Whiffen et al. 2007). The modified assay attenuates this interference by (i) subtracting the false positive signal and (ii) correcting for protein quenching from polyphenolic content — as described by Frølund et al. (1995). Using this approach, Redmile-Gordon et al. (2015) found that colorimetric estimations of EPS-protein were corroborated by more laborious gas chromatography of N-acetyl, O-isopropyl derivatives of hydrolysed constituent amino acids.
Microbial community composition
Extraction and purification of total community DNA
Based on the microbial EPS protein data, the highest influence was found for 10% clay addition. Therefore, we selected the two most contrasting clay treatments (i.e. 0% vs. 10%) for the analysis of the microbial community composition. Total community DNA (TC-DNA) was extracted from 0.5 g soil (wet weight) using the FastDNA®SPIN Kit for Soil (MP Biomedicals, Santa Ana, California) and purified with the GENECLEAN®SPIN Kit (MP Biomedicals, Santa Ana, California) following the instructions of the manufacturer. For cell lysis, a FastPrep™ FP120 (Qbiogene, Inc., Carlsbad, California) bead-beating system was used. The success of the extraction was checked by gel electrophoresis on a 0.8% agarose gel that was stained with ethidium bromide (0.005%) and photographed under UV-light (Intas Gel jet Imager 2004, Intas, Göttingen, Germany). Differences in DNA extraction efficiency cannot be fully excluded. Therefore, we focused our analysis on qualitative comparisons using the 16S rRNA gene-based community composition with relative proportion of abundant populations being less affected by total DNA amounts.
Denaturing gradient gel electrophoresis (DGGE) analysis of 16S rRNA gene amplicons
Purified TC-DNA served as a template for PCR with the primer pair F984-GC/R1378-1401 (Table S1) to amplify 16S rRNA gene fragments with a GC-clamp attached for DGGE analysis (Heuer et al. 1997; Nübel et al. 1996). One microlitre purified TC-DNA was added to a volume of 24 µl reaction mixture containing 1 × GoTaq® reaction buffer (Promega, Fitchburg, WI, US), 0.2 mM dNTPs, 3.75 mM MgCl2, 4% acetamide, 0.2 µM of each primer, and 0.05 U/µl GoTaq®Flexi polymerase. The reaction mixture was subjected to an initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 53 °C for 1 min, and elongation at 72 °C for 2 min and a final elongation step at 72 °C for 10 min. The presence of amplicons was checked by gel electrophoresis on a 1% agarose gel that was stained with ethidium bromide and photographed under UV light.
To fingerprint the soil microbial community composition, 16S rRNA gene amplicons were subjected to DGGE with the Ingeny PhorU system (Ingeny, Goes, the Netherlands) as described by Weinert et al. (2009). After electrophoresis at a constant voltage of 140 V for 17 h at 58 °C, the silver-staining procedure according to Heuer et al. (2001) was applied. After air-drying, the gel was scanned transmissively (Epson Perfection V700 Photo, Epson, Nagano, Japan).
Illumina sequencing of 16S rRNA gene amplicons
The hypervariable V3/V4 region of the 16S rRNA gene of the bacterial and archaeal kingdom was amplified by PCR using primers 341F (Sundberg et al. 2013) and 806R (Caporaso et al. 2011) as described by Chowdhury et al. (2019). The procedure of tagging, adding of sequencing adapters, purification, and high-throughput amplicon sequencing of 16S rRNA genes (2 × 250 bp, paired-end, Illumina MiSeq platform (Illumina, San Diego, CA, USA)), was applied as described by Nunes et al. (2016) and Chowdhury et al. (2019). Raw amplicon data were deposited at NCBI Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA748274. According to Nunes et al. (2016) sequences were demultiplexed and trimmed. Pairing and filtering (maxee 0.5) of the sequences were carried out using Usearch v7.0.1090 (Edgar 2010). Singletons were removed and sequences were clustered to operational taxonomic units (OTUs, sequence similarity cutoff 97%) with UPARSE (Edgar 2013). For chimaera filtering, Usearch and ChimeraSlayer package (Haas et al. 2011)) were used. The RDP16 database was used for classification of representative OTUs (cutoff 80%). Sequences that were unclassified at domain level or were affiliated to cyanobacteria/chloroplasts or mitochondria were removed from the data set, resulting in a total of 3,483 OTUs. On average, 12,150 quality-filtered sequences were obtained per sample.
Determination of aggregate size distribution and stability
To identify whether soil microorganisms foster aggregate formation, the aggregate size distribution was determined by dry sieving for each treatment and independent replicate. Dry soil material (40 g) was weighed into the sieving machine (AS 200 control “g”; Retsch, Haan, Germany), containing sieves 4 mm, 2 mm, 1 mm, and 0.25 mm. Pre-tests were conducted to obtain the most suitable sieving procedure to test aggregate size distribution with minimal disruption, especially in regard to the soil without clay addition. Accordingly, an amplitude of 0.5 mm for 1 min was selected for the determination of the aggregate size distribution. The sieved sample was collected separately into aggregate size fractions > 4 mm; 2–4 mm; 1–2 mm; 0.25–1 mm; and < 0.25 mm, and the mass of each size fraction was then determined. The aggregation size distribution index was calculated by the geometric mean diameter (GMD) as suggested index for aggregate size distributions (Kemper and Rosenau 1986; Larney 2008). The GMD was calculated using Eq. 2 according to Carmeis Filho et al. (2016):
where, wi is the weight of aggregates in the fraction i and di is the mean diameter of size fraction i.
The 1–2-mm aggregates were taken as the representative macroaggregate size fraction, and their stability was tested using the slow wetting test of Le Bissonnais (1996). Briefly, 1 g of aggregates was slowly wetted through the capillarity effect using a hanging water column technique. Afterwards, the soil material was carefully transferred to the wet sieving apparatus equipped with sieves of mesh 63 µm (Eijkelkamp Soil & Water, Giesbeek, Netherlands). The sieving was processed in ethanol for 3 min ± 5 s (stroke = 1.3 cm; frequency: 34 times min−1). The oven-dried aggregates > 63 µm were fractionated by dry sieving (AS 200 control “g”, Retsch, Haan, Germany) with an amplitude of 0.5 mm for 1 min using a combination of sieves: 2, 1, 0.60, 0.25, and 0.063 mm. The results were expressed as mean weight diameter (MWD) calculated according to Larney (2008) using Eq. 3:
where, mi is the weighted percentage of aggregates in the fraction i and di is the mean diameter of size fraction i.
Data analyses and statistics
The DGGE gel images were analysed with GelCompar II software version 6.6 (Applied Maths, Sint-Martens-Latem, Belgium). Based on Pearson’s correlation, the similarity coefficient values were calculated for each gel by pairwise comparison of the lanes. The resulting similarity matrices were used for cluster analysis by UPGMA algorithm.
A PERMANOVA (Bray–Curtis dissimilarity, 10,000 permutations) was conducted on transformed OTU data (number of reads relative to 100%) to compare microbial community compositions among samples differing in substrate and clay content on day 20 (beta-diversity). After data normalisation according to edgeR developer recommendations, likelihood-ratio tests under negative binominal distribution and generalised linear models were used to seek genera with significantly different (FDR-corrected p < 0.05) abundances depending on clay content or substrate type. Additionally, LefSe (linear discriminant analysis effect size) analysis (Segata et al. 2011) was performed based on relative abundance data to find discriminative OTUs that best explain differences in the microbial communities between the two clay contents (LDA > 4.0). Non-metric multidimensional scaling (NMDS; Bray–Curtis dissimilarity) was applied based on relative abundance OTU data to detect the effect of the treatments (with or without clay and substrate addition) on microbial community composition. The species richness as measure of alpha-diversity was calculated based on a 100 times randomly subsampled data set (n = 3933) and tested for the effect of substrate and clay content by two-way ANOVA (factors: substrate (levels: starch, cellulose), clay (levels: 0%, 10%)) as well as one-way ANOVA for the individual treatments followed by pairwise Tukey test comparisons (p < 0.05). For the microbial community, statistical analyses focused on the effect of substrate quality and clay amendment. The control treatment (no substrate without clay addition, day 0) was only used as reference and not included in the statistical analysis.
Generalised linear models were applied to determine the effects of substrate (starch or cellulose) addition and clay (0.1–10%) on aggregate formation (GMD), aggregate stability (MWD), EPS production, and microbial biomass C. The analyses were carried out for the entire period separately for each substrate. Differences in the magnitude of responses of all parameters (GMD, MWD, EPS, and microbial biomass C) to clay and substrate factors were analysed with Tukey HSD test at p < 0.05. Relationship of the soil aggregation parameters (GMD, MWD) with microbial parameters (EPS-protein, EPS-polysaccharide, respiration, and microbial biomass C) was evaluated using Spearman’s rank correlations. Furthermore, canonical correspondence analysis (CCA, 999 permutations) based on log transformed relative abundance OTU data was performed in order to investigate the relationship between the discriminative OTUs, the microbial community composition and EPS-protein, and the link to soil aggregation (MWD and GMD). Environmental variables and OTUs (p < 0.05) were fitted onto the ordination plot using the envfit function (999 permutations). All data analyses were conducted using R version 3.6.2, packages vegan, multcomp, questionr, agricolae, permute, edgeR, phyloseq, gplots, RColorBrewer, rioja, mvabund, plyr, phia, emmeans, dplyr, survival, ggplot2, and ggcorrplot (R Core Team 2019).
Results
Microbial biomass C and EPS
Generalised linear models (GLM) analyses indicated that both starch and cellulose addition had a significant effect on the microbial biomass. Starch and cellulose addition explained about 26% and 32% variation of the microbial biomass C, respectively. Clay addition explained < 5%, irrespective of the C quality. Incubation time explained about 61% of the variation with starch, and 29% with cellulose addition (Table 1). The microbial biomass increased rapidly following the addition of starch, but with cellulose, the increases were slow and steady up to day 10 (Fig. 2 and Fig. S1). The highest microbial biomass C was recorded at day 3 for starch and day 10 for cellulose. Thus, biomass C increased more rapidly with starch (easily degradable) than cellulose (more recalcitrant). There were no significant effects of clay addition, except where included at 10%, a reduction in total biomass C was evident at most time-points (Fig. 2 and Fig. S1). Respiration showed a similar pattern to biomass C (Fig. S2).
Microbial biomass C (MBC) represented with lines (n = 4 ± standard errors (SE)) and extracellular polymeric substances (EPS) protein (EPS-protein; columns), measured in the soil with or without substrate or clay (montmorillonite) addition (+ 0%, + 0.1%, + 1%, and + 10%) over the experimental period. Columns and line dots are mean ± SE of four replicates. Statistical comparisons are made between EPS-protein means within the same clay addition. Bars marked with # (decrease EPS-protein) or * (increase EPS-protein) are significantly different (p < 0.05) from the same treatment at day 0. For MBC significance, see Fig. S1
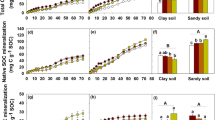
Microbial EPS production increased shortly after increases in biomass C and respiration. Among the experimental factors tested, GLM showed that EPS-protein and EPS-polysaccharide concentrations were explained primarily by clay addition (Table 1; 20–28%). EPS-protein was higher in soils with substrate amendment, and soils with additional clay (Fig. 2). EPS-protein in soils amended with starch peaked around day 3. This peak appeared at day 10 when 1% clay was added, and at day 40 with 10% additional clay (Fig. 2). In soils with 10% clay addition, the maximum EPS-protein concentration was measured at day 40 for both substrates (Fig. 2). Cellulose showed a similar pattern; except at 1% clay, the peak EPS-protein occurred a little later than with starch (day 20 vs. day 10). EPS-polysaccharide concentrations peaked at day 10 in response to starch, except with the highest clay addition, where the peak was recorded only at day 40 (Fig. 3). EPS-protein and polysaccharide concentrations in soils amended with substrates decreased substantially after day 10 unless they had also been amended with 10% clay. In summary, substrate addition resulted in increased EPS only if combined with clay addition. At high clay addition, starch resulted in higher EPS concentrations than cellulose (Figs. 2 and 3).
Microbial biomass C (MBC) represented with lines (n = 4 ± standard errors (SE)) with extracellular polymeric substances (EPS) polysaccharide (EPS-polysaccharide; columns), measured in the soil with or without substrate or clay (montmorillonite) addition (+ 0%, + 0.1%, + 1%, and + 10%) over the experimental period. Columns are mean ± SE of four replicates. Comparisons are made between EPS-polysaccharide means within the same clay addition. Bars marked with # (decrease EPS-polysaccharide) or * (increase EPS-polysaccharide) are significantly different (p < 0.05) from the same treatment at day 0. For MBC significance, see Fig. S1
Microbial community composition as influenced by clay content and substrate
Based on the DGGE result, there were no noticeable treatment-dependent shifts in the bacterial community on day 0 and day 3 (Fig. S1). Irrespective of the amount of added clay, strong shifts in the community composition due to the different types of substrate added (cellulose vs. starch) were observed after 10 and 20 days (Figs. S3–6). Thus, the two levels of clay treatments (+ 0 and + 10%) and substrate quality (cellulose and starch) sampled at day 20 were analysed by Illumina sequencing. Rarefaction curve analysis showed that sequencing depth was sufficient to cover diversity in all soil samples (Fig. S7). In addition, the treatment without substrate and clay amendment at day 0 was included as reference since DGGE analysis showed that microbial communities in soils without substrate addition did not change strongly over the incubation time (Figs. S3 and S6). Acidobacteria, Actinobacteria, Bacteroidetes, Candidatus Saccharibacteria, Chloroflexi, Firmicutes, Gemmatimonadetes, Proteobacteria, Thaumarchaeota, and Verrucomicrobia were the most dominant phyla (relative abundance higher than 1%) among all samples (Fig. 4, Table S2). Based on the analyses at phylum level, the composition of the microbial community was influenced by the added substrate. For example, the relative abundance of Firmicutes increased strongly (ca. fourfold) after addition of starch (S_ + 0%_20d). With clay addition, some phyla increased significantly in comparison to soils with either starch or cellulose and no clay addition, e.g. Acidobacteria (starch: ca. threefold, cellulose: ca. twofold), Deltaproteobacteria (starch: ca. twofold, cellulose: ca. 3.8-fold), and Alphaproteobacteria (cellulose:ca. 1.3-fold). The most prominent bacterial phyla, Bacteroidetes (highest relative abundance in soil amended with cellulose without clay addition) and Firmicutes (highest relative abundance in soil amended with starch without clay addition) showed a decrease in relative abundance (Bacteroidetes: − 15%, Firmicutes: − 50%) (Fig. 4; Table S2). The species richness of the microbial communities (Fig. S8) was slightly higher in soil with 10% clay added compared to soil without additional clay.
Mean (n = 4) relative abundance of microbial phyla and proteobacterial classes (> 1%) in the soil with and without clay or substrate addition (control: no added substrate (at day 0), C: cellulose addition, S: starch addition, + 0%: no clay addition, + 10%: 10% clay addition). Analyses are based on Illumina sequencing of 16S rRNA genes
According to the analysis of the taxonomic composition of the microbial communities at genus level, there were several genera among the most abundant that were significantly enriched in response to either clay content or substrate quality (Fig. S9; Tables 2 and 3). Within these genera, the strongest responders to cellulose addition (no clay addition) belonged to Sporocytophaga (Fig. S9; Table 3). For samples amended with starch (no clay addition), Paenibacillus was the most abundant genus. Comparison of soils amended with cellulose or starch at the two clay levels showed that the type and abundance of microbial genera varied with clay content. In the presence of 10% additional clay, the genus Sporocytophaga was rarely found in soil amended with cellulose, while the relative abundance of unclassified sequences within Chitinophagaceae and Myxococcales thrived in response to the 10% clay addition. In soils amended with starch, Herpetosiphon (phylum Chloroflexi) became the predominant genus (ca. 15.4%) increasing with clay addition (Fig. S9; Table 2).
The PERMANOVA analysis revealed that clay addition had the greater impact on microbial community composition. About 45% (p < 0.01) of variation in the OTU data set were explained by clay content, ca. 21% (p < 0.01) by substrate quality, and ca. 16% (p < 0.01) by the interaction of both factors (Table 4). The NMDS analyses confirmed that the clay content influenced the microbial community (Fig. S10). The NMDS showed clear separation of samples according to clay content along the NMDS1 axis. The community composition of samples amended with starch or cellulose were also separated along NMDS axis 2, and this effect was stronger without clay (+ 0%) than with clay. The microbial communities in the reference soil (without substrate or clay addition at day 0) were more similar to the soil without clay addition as shown along NMDS1 axis. Overall, the amount of clay in soil had a greater influence on the community composition than either substrate addition or substrate quality.
Formation and structural stability of soil aggregates
Effect of substrates — C quality/availability and clay addition
Geometric mean diameter (GMD) was used as an indicator of the extent of aggregate formation. Aggregate formation was mostly explained (92–93%) by the amount of clay added for all days, irrespective of the quality of the C source (Table 5). However, there was no significant difference in the aggregate size distribution and GMD among the soils with lower amounts of clay (+ 0%, + 0.1, and + 1%), with significant differences only observed with the addition of clay at 10% (Fig. 5). The proportion of aggregates < 250 µm decreased with increasing clay content, while the proportions > 4 mm and 2–4 mm were the highest in soils with 10% clay addition (Fig. 6). GLM analysis (Table 5) shows no significant effect of the added substrates, starch or cellulose, on measures of aggregate formation. To isolate the effect of substrate addition, a separate GLM analysis was considered excluding soils with clay added (Table 5). GLM showed no significant effect of the added substrate on aggregate formation, irrespective of the C source (Table 5). However, GMD was the highest (Fig. 5) for the soil with starch addition at day 3 (fast response) and soil with cellulose was the highest at day 40 (slow response). The mass of large aggregates (> 4 mm) in soils without clay addition was the highest at day 3 when microorganisms metabolised starch, and at day 40 when microorganisms metabolised cellulose, decreasing thereafter (Fig. 6).
Geometric mean diameter (GMD) of soil aggregates (a) with (0.1%, 1%, 10%) and without (0%) clay (montmorillonite) addition at each sampling day over the experimental period. Columns are mean ± standard errors (SE) of four replicates. Comparisons are made between GMD means within the same clay addition. Bars marked with * (increase in GMD) are significantly different (p < 0.05) from the same treatment at day 0
Soil aggregate size distributions of all treatment combinations over the experimental period; clay additions (%) are given for each substrate, and time-sequence (days 0–80); n = 4. Mean comparisons are presented only for the aggregate size class > 4 mm within the same levels of clay addition and substrate type. * denotes a statistically significant (p < 0.05) increase in aggregation vs. day 0
Mean weight diameter (MWD) of aggregate class 1–2 mm was used to investigate the structural stability of the aggregates (Fig. 7). Overall, the clay content (%) explained most of the variation observed in MWD during incubation: MWD increased slightly with clay content (Table 5). This was most significant for soils without substrate addition (Fig. 7). Similarly, the MWD increased to a greater extent by starch addition when no clay was added (Fig. 7). Substrate addition explained only about 31% of the variation observed in MWD when clay additions were excluded (Table 5). However, 3% was explained by substrate addition when all treatments were considered (Table 5). Soils amended with starch showed declines in aggregate stability after day 40. This decrease became less apparent with increasing clay content (Fig. 7).
Mean weight diameter (MWD) after stability test of the soil aggregate size fraction 1–2 mm. Columns represent means ± standard errors (SE) of four replicates. Bars marked with * (increase in MWD) are different vs. day 0 (within the same clay treatment) to a statistically significant margin (p < 0.05). Significantly different (p < 0.05) responses to clay addition (vs. + 0%) are indicated by ↑ (increase) and ↓ (decrease) for starch and ^ (increase) for control. Mean comparisons are presented among treatments in the same group (control and starch)
Relationship between the soil aggregation and microbial parameters
Overall, the Spearman’s rank correlations (Fig. 8a) indicated weak or no significant relationships between aggregate formation (GMD) and stability (MWD) and most of the microbial parameters — except for EPS. GMD was significantly positively correlated with the EPS-protein generated from added starch (0.58) and cellulose (0.41) (Fig. 8a). EPS-polysaccharide contents were negatively correlated with GMD in soil with no substrate or cellulose addition (Fig. 8a), and no significant correlation with EPS-polysaccharide was found in the soil with added starch (Fig. 8a). EPS-protein also showed a positive correlation with aggregate stability (MWD) in the soil without (0.35; Fig. 8a) and with substrate addition (0.34, Fig. 8a), respectively. There was no significant correlation of MWD with other microbial parameters (Fig. 8a).
Spearman’s rank correlation showing the relationship of geometric mean diameter (GMD) and the stability of the aggregate size fraction 1–2 mm (MWD with the microbial parameters (respiration (CO2), microbial biomass C, extracellular polymeric substances protein (EPS-protein and -polysaccharide) in a all soils and b soils without clay addition. Correlation coefficients marked with “X” are not significant (p < 0.05)
We also applied Spearman’s rank correlations solely to the soil without clay addition to evaluate the influence of microbial metabolism of substrate alone. When this was done, the GMD correlated with all other microbial parameters in soil with added starch, except for EPS-polysaccharide content (Fig. 8b). In contrast, for soil amended with cellulose (Fig. 8b), there was no strong correlation of the GMD with any of the microbial parameters, except for EPS-polysaccharide content. When clay was excluded, MWD showed negative correlations with all the microbial properties in the soil with no added substrate (Fig. 8b) and no significant correlations in the soil with added starch.
The CCA revealed a relationship between microbial community composition, EPS, and soil aggregation (Fig. 9). Ordination showed that the clay content-dependent separation of the microbial communities along the CCA1 axis, explaining 21.1% of the variance in the dataset, was clearly related to the amount of EPS-protein (Fig. 9a). The analysis revealed that the OTUs 383 (Thermomarinilinea lacunifontana), 633 (Aggregatilinea lenta), 210 (Flavitalea antarctica), 669 (Ornatilinea apprima), and 1419 (Flavisolibacter galbus), which were more abundant in 10% clay content soils, were positively correlated with EPS-protein. The community composition of samples amended with starch and 10% clay (Fig. 9b) was positively associated to MWD, GMD, and EPS-protein resulting in clay content-dependent separation along CCA1 axis, explaining 55.4% of the variance in the dataset (Fig. 9b). This revealed that OTUs 669 (Ornatilinea apprima), 383 (Thermomarinilinea lacunifontana), 210 (Flavitalea Antarctica), and 633 (Aggregatilinea lenta) were in addition also positively correlated to GMD. Detailed information about the discriminative OTUs is given in Table S3.
Canonical correspondence analysis (CCA) of microbial community compositions in soil samples. CCA is based on log10 transformed relative abundances of microbial OTUs. a OTUs of “No substrate addition_ + 0% clay_d0”, “Cellulose_ + 0% clay_d20”, “Starch_ + 0% clay_d20”, “Cellulose_ + 10% clay_d20","Starch_ + 10% clay_d20” treatments using EPS_protein as constraint variable. b OTUs of “No substrate addition_ + 0% clay_d0”, “Starch_ + 0% clay_d20","Starch_ + 10% clay_d20” treatments using EPS_protein, MWD and GMD as constraint variables. Significant vectors (p < 0.05) were fitted onto CCA ordinations including log10 transformed relative abundances of OTUs (Table S3) that showed a significantly different relative abundance among treatments with and without additional clay according to LefSe and edgeR anlaysis (a, OTU29; b, OTU383; c, OTU210; d, OTU1419; e, OTU669; f, OTU633; Cellulose, cellulose addition; Starch, starch addition; + 0% clay, no clay addition; + 10% clay, 10% clay addition; d, days)
Discussion
Microbial community response to clay and substrate additions
Decomposability of soil C is a central factor directly affecting (i) emergent microbial community composition (Goldfarb et al. 2011) and (ii) extracellular biochemical properties (Redmile-Gordon et al. 2015). These direct effects are complemented by contrasting biophysical feedback from alterations to the EPS matrix, further affecting the composition of the microbial community (Or et al. 2007). Accordingly, we expected contrasts in the responses of microbial communities to the added substrate (labile starch vs. more recalcitrant cellulose) and these were seen through shifts in the microbial biomass and community composition (Fig. 2). For instance, bacteria of the phylum Firmicutes responded most strongly to the addition of starch, while bacteria of the phylum Bacteroidetes responded most strongly to cellulose addition (Fig. 4). Bacteroidetes are known to degrade complex organic compounds and were previously reported as key degrader of cellulose in microbial community profiling (Doud et al. 2020). Bacteria belonging to the genus Paenibacillus (phylum Firmicutes) produce amylase, a key enzyme in the metabolism of starch (Grady et al. 2016; Rajesh et al. 2013). Clearly, responses to substrates are taxon-specific; hence, the community composition adapts accordingly. Contrasting microbial community responses to substrate addition have also been reported previously (Tan et al. 2019). However, we observed further that the addition of clay enabled increases in microbial biomass to occur in response to substrate without compromising on microbial diversity. Overall, we found in our study that while the microbial biomass increased in response to the substrate addition (Fig. 2 and Fig. S1), the composition of the microbial community varied most according to the soil clay content (Fig. 4).
The observed changes in the microbial community composition may be associated with physical habitat changes within the formed aggregates as affected by clay addition (10%; Fig. 6). For example, Philippot et al. (1996) showed a change in the distribution of a denitrifying bacteria in artificial aggregates owing to the development of an anoxic zone in the centre of aggregates a few weeks after incubation. Similarly, Chenu et al. (2001) reported a heterogeneous microbial distribution along the spatial axis in soil aggregates of sandy and clayey soil following glucose addition, with higher bacterial counts on the aggregate surfaces than the inner pores. They attributed this to inhibition of glucose penetration into the aggregates (Chenu et al. 2001). Such interruptions in substrate flow may be higher in clayey soil with smaller pores and greater occlusion vs. sandy soil.
In the literature, there are contrasting reports about the effect of clay minerals on soil microbial biomass. While some reports show a positive effect of clay (Amato and Ladd 1992; Rakhsh et al. 2020; Wei et al. 2014), there are others showing the reverse (Liddle et al. 2020). On the one hand, clay has been demonstrated to protect microbes against toxicity and predation (Babich and Stotzky 1977; McMahon et al. 2016; Rutherford and Juma 2004; Su et al. 2021). For example, Rutherford and Juma (2004) found that bacteria are more protected against predation in fine-textured (more clayey) than in coarse-textured soils. On the other hand, clay minerals (kaolinite and goethite) were found to cause mortality in Bacillus (Ma et al. 2017). Krause et al. (2019) found a decrease in the number of culturable Pseudomonas protegens cells in the presence of montmorillonite and goethite in an experiment involving the incubation of these bacteria with a mineral suspension. Furthermore, increasing clay contents were previously found to reduce microbial enzyme activities (Olagoke et al. 2019; Quiquampoix et al. 2002) and changes in the biogeochemistry of the microhabitat have been observed due to the composition of clay minerals (Babin et al. 2014; Cuadros 2018; Uroz et al. 2015). In our study, the relative abundance of some bacterial genera associated with the phyla Firmicutes (e.g. Paenibacillus, Cohnella), Proteobacteria (e.g. Legionella, Aquicella) and Bacteroidetes (e.g. Sporocytophaga, Tumebacillus) in response to starch or cellulose was less pronounced in the presence of added clay (Fig. 4 and Table 2). This finding aligns with Biswas et al. (2020) who observed a reduced relative abundance of Firmicutes and Proteobacteria following the addition of 5% bentonite (montmorillonite) to soil. Altogether, these studies confirm that clay minerals can suppress growth and activity — favouring less copiotrophic organisms.
Nevertheless, the high alpha-diversity with 10% clay addition and enrichment of previously less abundant genera with clay addition such as Sphingomonas, Gemmatimonas, Chitinophagaceae-unclassified, and Herpetosiphon gives an indication of niche differentiation being promoted under higher clay contents. This could also depend on variation in the metabolic responses of individual bacterial groups adhering to clay minerals (Żur et al. 2016). Our hypothesis that substrate quality (C quality) affects the composition of the microbial community was supported, but it was affected more by the clay content, which also enhanced microbial diversity. Our results confirm studies pointing out that physicochemical properties of clay minerals are a key driver of the microbial community composition in soil (Biesgen et al. 2020; Biswas et al. 2020; Uroz et al. 2015).
EPS production
Substrate addition promoted soil respiration (Fig. S2), indicating an increase in microbial activity, responding more rapidly to starch than cellulose. In the present study, analyses indicated that the production of EPS lagged behind growth and respiration, as is expected from the work of others who found EPS production to peak around the transition from the exponential growth phase to the stationary phase (Abid et al. 2018; Wingender et al. 1999). Where clay additions were 1% or less, EPS concentrations peaked at day 10 (Figs. 2 and 3). However, with the addition of clay at 10%, EPS production increased slowly and was sustained: peaking at day 40. Ma et al. (2017) suggested that EPS production was a tolerance mechanism to protect cells against physical harm from soil minerals. This explanation would seem to support the steady accumulation of EPS in our study responding to clay addition. Within sandy soils, added C typically metabolises at a rapid rate as it is not inhibited by lack of oxygen availability or occlusion by clay (Fichtner et al. 2019; Neira et al. 2015). Accordingly, microbial biomass C peaked earliest without additional clay (Fig. 2) and very little EPS-protein was detected after day 10 in soils not amended with additional clay (Fig. 2). Nevertheless, the higher production of EPS by microbes in the soil with a higher clay content as compared to the soil with no clay addition (Fig. 2) shows that there is a mechanism or a reaction process stimulating the microbes to produce EPS in response to increasing clay contents.
A number of studies have analysed clay-microbe relationships (Brennan et al. 2014; Krause et al. 2019; Ladd et al. 1996; Mueller 2015). On the one hand, microbes can induce weathering of minerals: affecting their chemical and physical properties (Hong et al. 2016). On the other hand, adsorption to clay minerals can reduce the activities of the extracellular enzymes produced by the microorganisms (Olagoke et al. 2019), thereby slowing nutrient acquisition. This may also ignite a metabolic response in microbes, although this could depend on stress tolerance (Serrazanetti et al. 2009; Żur et al. 2016). The production of EPS has been described as a strategy by which microorganisms protect themselves in extreme conditions (Costa et al. 2018). Variation in abiotic conditions such as drought, temperature, pH, and salinity can trigger the production of EPS as a response to environmental stresses (Sandhya and Ali 2015). Taking into account the inhibited microbial growth of microbes due to clay addition (Fig. 2 and Fig. S1) and presumed interference between extracellular enzymes and clay minerals, the concomitant increases in EPS production may help to keep the activity of extracellular enzymes high, as found in aquatic environments (Romaní et al. 2008). Our findings concur with those of Redmile-Gordon et al. (2015) where resource limitation (N) caused an increase in the quantity of EPS produced by the soil microbial biomass. These authors hypothesised that removal of the stress removed competitive advantage from the protection of enzymes within a (metabolically expensive) matrix of EPS. By studying genes related to the production of EPS, Cania et al. (2020) found a lower relative abundance of wza in a finer-textured soil, while other genes related to EPS production were maintained throughout soils of different textures. The authors assumed that lower relative abundance of this gene might indicate more favourable conditions and therefore lower EPS production. However, within the present study, we found that the additional clay caused slow and steady increases in EPS (Figs. 2 and 3). Accordingly, we speculate that the microorganisms invested proportionally more C into EPS when clay was increased, either to reduce deleterious impacts caused by the clays directly or because of some clay-induced limitation on resource acquisition.
Aggregate formation and stability
Role of added substrates vs. clay and microbial parameters
We observed major differences in the formation of aggregates related mostly to the addition of clay. Evidently, the contribution of the microbial biomass (and associated EPS) to aggregation is superimposed by the soil clay content. For example, the soil with clay addition showed no significant effect of microbial biomass C on aggregate formation (i.e. the GMD); there was only a significant effect when no clay was added (Fig. 8). Earlier studies have revealed different and also contradictory effects of microbial biomass C on soil aggregation. Bossuyt et al. (2001) pointed out a poor correlation of microbial biomass to soil aggregation, while other studies established a good correlation between microbial biomass C and soil aggregation (Andruschkewitsch et al. 2014; Kiem and Kandeler 1997). As shown by the CCA analyses, the abundance of some species was correlated with soil aggregation (Fig. 9b). For instance, among those positively correlated to GMD and EPS-protein were members of the family Anaerolinaceae (OTU 383, 669, 633, Fig. 9b, Table S3). This complements the findings of Cania et al. (2019) who suggested this family had a propensity to produce lipopolysaccharides. Our findings suggest a possible role for Anaerolinaceae in soil aggregation. In our study, members of this family were more abundant in the soil amended with 10% clay (Fig. S11). So, while the changes in aggregate properties may be due to specific taxa — their contributions were enabled by the addition of clay. The current findings illustrate the dominant influence of clay for aggregate formation, as determined by GMD. We suggest further studies on how substrate and clay additions induce changes in microbial communities and affect the formation of aggregates.
Our results signify that more readily available substrates such as starch induce a more rapid formation of soil aggregates than less bioavailable substrates such as cellulose. In soils with substrate but no clay addition, we found that the aggregate size fraction > 4 mm peaked at day 3 with starch and day 40 with cellulose, in each case decreasing thereafter (Fig. 6). This echoes in part the findings of Mizuta et al. (2015), where macroaggregates (> 2 and 1–2 mm) increased at the early stage of incubation (0–6 days) by the addition of starch and later decreased to the control level at the end of incubation (99 days). In contrast, these authors could not find a consistent trend in response to the addition of cellulose. In the present study, the increased proportion of macroaggregates (> 4 mm) in soil with starch addition coincided with an increase in respiration at day 3. This is an indication that the microbial decomposition of substrate to CO2 and intermediates followed by anabolic processes contributed to soil aggregate formation. Presumably, the conversion of a fraction of substrate-C into microbial EPS contributes to the formation of stable soil aggregates. The soil with starch addition showed greater aggregate stability compared to the soil with no substrate addition, which concurs with the work of others (Abiven et al. 2009; Andruschkewitsch et al. 2014). The soil with starch addition also showed the highest EPS production, especially when clay was added. The two species, Flavitalea antarctica (OTU 210) and Flavisolibacter galbus (OTU 1419), found to be correlated with aggregation and EPS-protein (Fig. 9, Table S3) were significantly abundant in the high clay soil (Table 3 and Table S3, Fig. S11). These species belong to the Chitinophagaceae family. This family was suggested by Cania et al. (2019) to be involved in enhanced EPS production. These interrelations indicate that microorganisms contribute to soil aggregation by producing EPS. Among the microbial parameters determined, EPS, especially EPS-protein, mostly correlated with soil aggregate formation but only weakly with aggregate stability. However, our result confirms the hypothesis that the quantity of EPS produced by microorganisms — and its composition — is important for aggregate formation.
EPS in soil aggregation and stability: EPS-protein vs. EPS-polysaccharide
Most functions attributed to EPS relate to protection of the producing microorganisms (Costa et al. 2018). Because of the cell-to-cell aggregation resulting from EPS production, it is assumed that EPS could form a glueing agent controlling aggregate formation and stability. As mentioned earlier, we found a significant correlation between EPS and soil aggregation. We noted differences in how the two major components of EPS, proteins and polysaccharide, varied with the addition of clay and their relation to aggregate formation and stability. While EPS-protein production seems to be enhanced by increasing clay content, EPS-polysaccharide was mostly lowest in the soil with the highest clay content (Fig. 3).
EPS-protein was significantly correlated with aggregate formation and stability, although weakly (Fig. 8). The correlation became stronger for aggregate formation with the addition of substrate. EPS production is an energy demanding process; thus, with the availability of substrate, the microorganisms could derive energy for that production. The soil with 10% clay addition had the highest GMD with the peak at day 40. The highest EPS-protein was also observed at day 40 in the same treatment. For the EPS-polysaccharide, no consistent pattern was found. In contrast to some studies on the role of polysaccharide for soil aggregation (de Caire et al. 1997; Harahap et al. 2018; Zethof et al. 2020), EPS-polysaccharide showed weak and negative or no correlation with soil aggregate formation without clay addition (Fig. 8b). These findings support those of Redmile-Gordon et al. (2020) who found EPS-protein concentrations were more closely related to aggregate stability than EPS-polysaccharide. Polysaccharides vary in their composition and functions (Holtekjølen et al. 2006; Singh et al. 2019). Some have protective roles, e.g. alginate (Limoli et al. 2015), others have good gelling/structural properties such as adhesion as gellan and dextran (Freitas et al. 2011; Limoli et al. 2015; Wilkinson 1958). While it is clear that not all EPS-polysaccharides contribute to aggregate formation and stability, the same may also be true for EPS-protein. To our knowledge, enzymes of the EPS matrix have no role in physical stabilisation. However, biofilm structural proteins are known to self-assemble extracellularly: those of Bacillus subtilis for example providing stability and protecting fragile polysaccharides against dispersal (Arnaouteli et al. 2016). Redmile-Gordon et al. (2020) proposed that concomitant increases in EPS-protein and soil aggregate stability were due to the synthesis of EPS rich in hydrophobic amino acids — previously observed to increase in the same soil when given a hydrophilic source of C (Redmile-Gordon et al. 2015). Biologically induced hydrophobicity is known to increase the stability of soils by lowering the wettability and increasing the cohesion of aggregates (Chenu et al. 2000).
Besides showing that proteins in the EPS appear to be more important for soil aggregation than polysaccharide, we found clear signs of EPS decomposition also occurring after day 10. This novel finding demonstrates that the EPS itself is also a relatively labile component of soil organic matter — especially in the sandy soil (Figs. 2 and 3). The higher EPS-protein concentrations present in response to clay may be due to some stabilising effect against their decomposition as clay minerals are acknowledged to protect organic compound against degradation by decomposers (McMahon et al. 2016). It is perhaps owing to this that the large additions of clay (to 10% mass) were more pivotal for aggregate stability than small variations in EPS-protein over all of the treatment combinations. This also lends support to the hypothesis of De Gryze et al. (2006) that aggregate formation is induced mainly by glueing effects of EPS, and that texture affects aggregation more during later stages of the aggregate lifetime than during aggregate formation.
Conclusion
Our study showed that substrate quality affects the community composition and population dynamics of microbes in soil. However, the microbial responses to substrate and effects on soil aggregation depended more strongly on the variation in the environmental conditions related to the clay content. Overall, we found that without clay addition, increased microbial activity by amendment with labile OM did not result in greater EPS production. While substrate addition supported microbial growth, soil texture had a stronger effect on the divergence of microbial community composition and microbial production of EPS. Nevertheless, we found that EPS-protein produced by soil microbes contributed to soil aggregate formation, but soil aggregate stability depended more on the heterogeneity in soil texture. As such, increasing clay content enhanced aggregate stability and additionally resulted in the development of distinct microbial communities and increased quantities of proteinaceous EPS. Our mechanistic findings in this model system support results in soils taken from the field where proteinaceous moieties were more closely correlated to soil aggregate stability than EPS polysaccharides (Redmile-Gordon et al. 2020). We suggest further research to address microbe-clay mineral interactions in order to disentangle the mechanisms responsible for enhanced microbial production of EPS discovered with increasing clay contents.
References
Abid Y, Casillo A, Gharsallah H, Joulak I, Lanzetta R, Corsaro MM, Attia H, Azabou S (2018) Production and structural characterization of exopolysaccharides from newly isolated probiotic lactic acid bacteria. Int J Biol Macromol 108:719–728
Abiven S, Menasseri S, Chenu C (2009) The effects of organic inputs over time on soil aggregate stability–a literature analysis. Soil Biol Biochem 41:1–12
Amato M, Ladd JN (1992) Decomposition of 14C-labelled glucose and legume material in soils: properties influencing the accumulation of organic residue C and microbial biomass C. Soil Biol Biochem 24:455–464. https://doi.org/10.1016/0038-0717(92)90208-F
Andruschkewitsch R, Geisseler D, Dultz S, Joergensen R-G, Ludwig B (2014) Rate of soil-aggregate formation under different organic matter amendments—a short-term incubation experiment. J Plant Nutr Soil Sci 177:297–306. https://doi.org/10.1002/jpln.201200628
Arnaouteli S, MacPhee CE, Stanley-Wall NR (2016) Just in case it rains: building a hydrophobic biofilm the Bacillus subtilis way. Curr Opin Microbiol 34:7–12. https://doi.org/10.1016/j.mib.2016.07.012
Babich H, Stotzky G (1977) Reductions in the toxicity of cadmium to microorganisms by clay minerals. Appl Environ Microbiol 33:696–705. https://doi.org/10.1128/aem.33.3.696-705.1977
Babin D, Vogel C, Zühlke S, Schloter M, Pronk GJ, Heister K, Spiteller M, Kögel-Knabner I, Smalla K (2014) Soil mineral composition matters: response of microbial communities to phenanthrene and plant litter addition in long-term matured artificial soils. PLoS ONE 9:e106865–e106865. https://doi.org/10.1371/journal.pone.0106865
Biesgen D, Frindte K, Maarastawi S, Knief C (2020) Clay content modulates differences in bacterial community structure in soil aggregates of different size. Geoderma 376:114544. https://doi.org/10.1016/j.geoderma.2020.114544
Biswas B, Juhasz AL, Mahmudur Rahman M, Naidu R (2020) Modified clays alter diversity and respiration profile of microorganisms in long-term hydrocarbon and metal co-contaminated soil. Microb Biotechnol 13:522–534. https://doi.org/10.1111/1751-7915.13510
Bossuyt H, Denef K, Six J, Frey S, Merckx R, Paustian K (2001) Influence of microbial populations and residue quality on aggregate stability. Appl Soil Ecol 16:195–208
Brennan FP, Moynihan E, Griffiths BS, Hillier S, Owen J, Pendlowski H, Avery LM (2014) Clay mineral type effect on bacterial enteropathogen survival in soil. Sci Total Environ 468:302–305
Cania B, Vestergaard G, Krauss M, Fliessbach A, Schloter M, Schulz S (2019) A long-term field experiment demonstrates the influence of tillage on the bacterial potential to produce soil structure-stabilizing agents such as exopolysaccharides and lipopolysaccharides. Environ Microbiome 14:1. https://doi.org/10.1186/s40793-019-0341-7
Cania B, Vestergaard G, Suhadolc M, Mihelič R, Krauss M, Fliessbach A, Mäder P, Szumełda A, Schloter M, Schulz S (2020) Site-specific conditions change the response of bacterial producers of soil structure-stabilizing agents such as Exopolysaccharides and Lipopolysaccharides to tillage intensity. Front Microbiol 11. https://doi.org/10.3389/fmicb.2020.00568
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS 108:4516–4522. https://doi.org/10.1073/pnas.1000080107
CarmeisFilho AC, Crusciol CA, Guimarães TM, Calonego JC, Mooney SJ (2016) Impact of amendments on the physical properties of soil under tropical long-term no till conditions. PLoS One 11:e0167564
Chenu C, Le Bissonnais Y, Arrouays D (2000) Organic matter influence on clay wettability and soil aggregate stability. Soil Sci Soc Am J 64:1479–1486
Chenu C, Hassink J, Bloem J (2001) Short-term changes in the spatial distribution of microorganisms in soil aggregates as affected by glucose addition. Biol Fertility Soils 34:349–356
Chenu C, Cosentino D (2011) Microbial regulation of soil structural dynamics. In: K Ritz, I Young (Eds) The Architecture and Biology of Soils: Life in Inner Space. CAB International Wallingford, UK, pp 33–70.
Chowdhury SP, Babin D, Sandmann M, Jacquiod S, Sommermann L, Sørensen SJ, Fliessbach A, Mäder P, Geistlinger J, Smalla K (2019) Effect of long-term organic and mineral fertilization strategies on rhizosphere microbiota assemblage and performance of lettuce. Environ Microbiol 21:2426–2439
Costa OYA, Raaijmakers JM, Kuramae EE (2018) Microbial extracellular polymeric substances: ecological function and impact on soil aggregation. Front Microbiol 9. https://doi.org/10.3389/fmicb.2018.01636
Cuadros J (2018) Clay minerals interaction with microorganisms: a review. Clay Miner 52:235–261. https://doi.org/10.1180/claymin.2017.052.2.05
de Caire GZ, De Cano MS, De Mule MZ, Palma R, Colombo K (1997) Exopolysaccharide of Nostoc muscorum (Cyanobacteria) in the aggregation of soil particles. J Appl Phycol 9:249–253
De Gryze S, Six J, Brits C, Merckx R (2005) A quantification of short-term macroaggregate dynamics: influences of wheat residue input and texture. Soil Biol Biochem 37:55–66. https://doi.org/10.1016/j.soilbio.2004.07.024
De Gryze S, Jassogne L, Bossuyt H, Six J, Merckx R (2006) Water repellence and soil aggregate dynamics in a loamy grassland soil as affected by texture. Eur J Soil Sci 57:235–246. https://doi.org/10.1111/j.1365-2389.2005.00733.x
Doetterl S, Berhe AA, Nadeu E, Wang Z, Sommer M, Fiener P (2016) Erosion, deposition and soil carbon: A review of process-level controls, experimental tools and models to address C cycling in dynamic landscapes. Earth-Sci Rev 154:102–122. https://doi.org/10.1016/j.earscirev.2015.12.005
Doud DFR, Bowers RM, Schulz F, De Raad M, Deng K, Tarver A, Glasgow E, Vander Meulen K, Fox B, Deutsch S, Yoshikuni Y, Northen T, Hedlund BP, Singer SW, Ivanova N, Woyke T (2020) Function-driven single-cell genomics uncovers cellulose-degrading bacteria from the rare biosphere. ISME J 14:659–675. https://doi.org/10.1038/s41396-019-0557-y
DuBois M, Gilles KA, Hamilton JK, Rebers PA, Smith F (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356. https://doi.org/10.1021/ac60111a017
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. J Bioinform 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Fichtner T, Goersmeyer N, Stefan C (2019) Influence of soil pore system properties on the degradation rates of organic substances during soil aquifer treatment (SAT). Appl Sci 9:496
Freitas F, Alves VD, Reis MA (2011) Advances in bacterial exopolysaccharides: from production to biotechnological applications. Trends Biotechnol 29:388–398
Frølund B, Griebe T, Nielsen PH (1995) Enzymatic activity in the activated-sludge floc matrix. Appl Microbiol Biotechnol 43:755–761. https://doi.org/10.1007/BF00164784
Frølund B, Palmgren R, Keiding K, Nielsen PH (1996) Extraction of extracellular polymers from activated sludge using a cation exchange resin. Water Res 30:1749–1758. https://doi.org/10.1016/0043-1354(95)00323-1
Goldfarb K, Karaoz U, Hanson C, Santee C, Bradford M, Treseder K, Wallenstein M, Brodie E (2011) Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front Microbiol 2. https://doi.org/10.3389/fmicb.2011.00094
Grady EN, MacDonald J, Liu L, Richman A, Yuan Z-C (2016) Current knowledge and perspectives of Paenibacillus: a review. Microb Cell Factories 15:203–203. https://doi.org/10.1186/s12934-016-0603-7
Guggenberger G, Elliott ET, Frey SD, Six J, Paustian K (1999) Microbial contributions to the aggregation of a cultivated grassland soil amended with starch. Soil Biol Biochem 31:407–419. https://doi.org/10.1016/S0038-0717(98)00143-6
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Petrosino JF, Knight R, Birren BW (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. https://doi.org/10.1101/gr.112730.110
Harahap N, Dwi AS, Gofar N (2018) The potential of exopolysaccharide-producing bacteria from rhizosphere of rubber plants for improving soil aggregate. J Degrade Min Land Manage 5:1275
Helfrich M, Ludwig B, Thoms C, Gleixner G, Flessa H (2015) The role of soil fungi and bacteria in plant litter decomposition and macroaggregate formation determined using phospholipid fatty acids. Appl Soil Ecol 96:261–264. https://doi.org/10.1016/j.apsoil.2015.08.023
Heuer H, Krsek M, Baker P, Smalla K, Wellington EM (1997) Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol 63:3233–3241. https://doi.org/10.1128/AEM.63.8.3233-3241.1997
Heuer H, Wieland J, Schönfeld J, Schönwälder A, Gomes NCM, Smalla K (2001) Bacterial community profiling using DGGE or TGGE analysis. In: Rochelle PA (ed) Environmental molecular microbiology: protocols and applications. Horizon Scientific Press, Wymondham, UK, pp 177–190
Holtekjølen AK, Uhlen AK, Bråthen E, Sahlstrøm S, Knutsen SH (2006) Contents of starch and non-starch polysaccharides in barley varieties of different origin. Food Chem 94:348–358. https://doi.org/10.1016/j.foodchem.2004.11.022
Hong H, Fang Q, Cheng L, Wang C, Churchman GJ (2016) Microorganism-induced weathering of clay minerals in a hydromorphic soil. Geochim Cosmochim Acta 184:272–288
Jenkinson DS, Brookes PC, Powlson DS (2004) Measuring soil microbial biomass. Soil Biol Biochem 36:5–7. https://doi.org/10.1016/j.soilbio.2003.10.002
Kaiser M, Ellerbrock RH, Sommer M (2009) Separation of coarse organic particles from bulk surface soil samples by electrostatic attraction. Soil Sci Soc Am J 73:2118–2130. https://doi.org/10.2136/sssaj2009.0046
Kemper WD, Rosenau RC (1986) Aggregate stability and size distribution. In: Arnold K (ed) Methods of Soil Analysis. Soil Sceince Society of America, Madison, Wisconsin, pp 425–442
Kiem R, Kandeler E (1997) Stabilization of aggregates by the microbial biomass as affected by soil texture and type. Appl Soil Ecol 5:221–230. https://doi.org/10.1016/S0929-1393(96)00132-1
Krause L, Biesgen D, Treder A, Schweizer SA, Klumpp E, Knief C, Siebers N (2019) Initial microaggregate formation: association of microorganisms to montmorillonite-goethite aggregates under wetting and drying cycles. Geoderma 351:250–260
Ladd JN, Foster R, Nannipieri P, Oades J (1996) Soil structure and biological activity. In: Stotzky G, Bollag J-M (eds) Soil biochemistry. Marcel Dekker Inc, New York, pp 23–78
Larney F (2008) Dry-aggregate size distribution. In: Gregorich EG, Carter MR (eds) Soil sampling and methods of analysis. Canadian Society of Soil Science, Boca Raton, Florida, USA, pp 821–831
Le Bissonnais YI (1996) Aggregate stability and assessment of soil crustability and erodibility: I. Theory and methodology. Eur J Soil Sci 47:425–437
Lehmann J, Kleber M (2015) The contentious nature of soil organic matter. Nature 528:60–68. https://doi.org/10.1038/nature16069
Liddle K, McGonigle T, Koiter A (2020) Microbe biomass in relation to organic carbon and clay in soil. Soil Syst 4:41
Limoli DH, Jones CJ, Wozniak DJ (2015) Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol spectr 3: https://doi.org/10.1128/microbiolspec.MB-0011-2014
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Ma W, Peng D, Walker SL, Cao B, Gao C-H, Huang Q, Cai P (2017) Bacillus subtilis biofilm development in the presence of soil clay minerals and iron oxides. npj Biofilms & Microbiomes 3: 4. https://doi.org/10.1038/s41522-017-0013-6
Martin JP, Waksman SA (1940) Influence of microorganisms on soil aggregation and erosion. Soil Sci 50:29–48
McMahon S, Anderson RP, Saupe EE, Briggs DEG (2016) Experimental evidence that clay inhibits bacterial decomposers: implications for preservation of organic fossils. Geology 44:867–870. https://doi.org/10.1130/g38454.1
Mizuta K, Taguchi S, Sato S (2015) Soil aggregate formation and stability induced by starch and cellulose. Soil Biol Biochem 87:90–96
Mueller B (2015) Experimental interactions between clay minerals and bacteria: a review. Pedosphere 25:799–810. https://doi.org/10.1016/S1002-0160(15)30061-8
Neira J, Ortiz M, Morales L, Acevedo E (2015) Oxygen diffusion in soils: understanding the factors and processes needed for modeling. Chil J Agric Res 75:35–44
Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann RI, Ludwig W, Backhaus H (1996) Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol 178:5636–5643. https://doi.org/10.1128/jb.178.19.5636-5643.1996
Nunes I, Jacquiod S, Brejnrod A, Holm PE, Johansen A, Brandt KK, Priemé A, Sørensen SJ (2016) Coping with copper: legacy effect of copper on potential activity of soil bacteria following a century of exposure. FEMS Microbiol Ecol 92. https://doi.org/10.1093/femsec/fiw175
Oades JM (1984) Soil organic matter and structural stability: mechanisms and implications for management. Plant Soil 76:319–337. https://doi.org/10.1007/bf02205590
Olagoke FK, Kalbitz K, Vogel C (2019) Control of soil extracellular enzyme activities by clay minerals—perspectives on microbial responses. Soil Syst 3:64. https://doi.org/10.3390/soilsystems3040064
Olagoke FK, Kaiser K, Mikutta R, Kalbitz K, Vogel C (2020) Persistent activities of extracellular enzymes adsorbed to soil minerals. Microorganisms 8:1796. https://doi.org/10.3390/microorganisms8111796
Or D, Phutane S, Dechesne A (2007) Extracellular polymeric substances affecting pore-scale hydrologic conditions for bacterial activity in unsaturated soils. Vadose Zone J 6:298–305
Philippot L, Renault P, Sierra J, Hénault C, Clays-Josserand A, Chenu C, Chaussod R, Lensi R (1996) Dissimilatory nitrite-reductase provides a competitive advantage to Pseudomonas sp. RTC01 to colonise the centre of soil aggregates. FEMS Microbiol Ecol 21:175–185. https://doi.org/10.1111/j.1574-6941.1996.tb00345.x
Quiquampoix H, Servagent-Noinville S, Baron M-H (2002) Enzyme adsorption on soil mineral surfaces and consequences for the catalytic activity. In: Dick RP (ed) RG Burns. Enzymes in the environment Marcel Dekker, New York, pp 285–306
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing.
Rajesh T, Kim YH, Choi Y-K, Jeon JM, Kim HJ, Park S-H, Park H-Y, Choi K-Y, Kim H, Kim HJ, Lee SH, Yang Y-H (2013) Identification and functional characterization of an α-amylase with broad temperature and pH stability from Paenibacillus sp. Appl Biochem Biotechnol 170:359–369. https://doi.org/10.1007/s12010-013-0197-z
Rakhsh F, Golchin A, Beheshti Al Agha A, Nelson PN (2020) Mineralization of organic carbon and formation of microbial biomass in soil: effects of clay content and composition and the mechanisms involved. Soil Biol Biochem 151:108036. https://doi.org/10.1016/j.soilbio.2020.108036
Redmile-Gordon MA, Armenise E, White RP, Hirsch PR, Goulding KWT (2013) A comparison of two colorimetric assays, based upon Lowry and Bradford techniques, to estimate total protein in soil extracts. Soil Biol Biochem 67:166–173. https://doi.org/10.1016/j.soilbio.2013.08.017
Redmile-Gordon MA, Brookes PC, Evershed RP, Goulding KWT, Hirsch PR (2014) Measuring the soil-microbial interface: extraction of extracellular polymeric substances (EPS) from soil biofilms. Soil Biol Biochem 72:163–171. https://doi.org/10.1016/j.soilbio.2014.01.025
Redmile-Gordon MA, Evershed RP, Hirsch PR, White RP, Goulding KWT (2015) Soil organic matter and the extracellular microbial matrix show contrasting responses to C and N availability. Soil Biol Biochem 88:257–267. https://doi.org/10.1016/j.soilbio.2015.05.025
Redmile-Gordon M, Gregory AS, White RP, Watts CW (2020) Soil organic carbon, extracellular polymeric substances (EPS), and soil structural stability as affected by previous and current land-use. Geoderma 363:114143. https://doi.org/10.1016/j.geoderma.2019.114143
Romaní AM, Fund K, Artigas J, Schwartz T, Sabater S, Obst U (2008) Relevance of polymeric matrix enzymes during biofilm formation. Microb Ecol 56:427–436
Rutherford PM, Juma NG (2004) Influence of texture on habitable pore space and bacterial-protozoan populations in soil. Biol Fertility Soils 12:221–227
Sandhya V, Ali SZ (2015) The production of exopolysaccharide by Pseudomonas putida GAP-P45 under various abiotic stress conditions and its role in soil aggregation. Microbiology 84:512–519. https://doi.org/10.1134/S0026261715040153
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. https://doi.org/10.1186/gb-2011-12-6-r60
Serrazanetti DI, Guerzoni ME, Corsetti A, Vogel R (2009) Metabolic impact and potential exploitation of the stress reactions in lactobacilli. Food Microbiol 26:700–711. https://doi.org/10.1016/j.fm.2009.07.007
Sheng GP, Yu HQ, Yue Z (2006) Factors influencing the production of extracellular polymeric substances by Rhodopseudomonas acidophila. Int Biodeterior Biodegrad 58:89–93. https://doi.org/10.1016/j.ibiod.2006.07.005
Singh JK, Adams FG, Brown MH (2019) Diversity and function of capsular polysaccharide in Acinetobacter baumannii. Front Microbiol 9. https://doi.org/10.3389/fmicb.2018.03301
Six J, Bossuyt H, Degryze S, Denef K (2004) A history of research on the link between (micro)aggregates, soil biota, and soil organic matter dynamics. Soil Tillage Res 79:7–31. https://doi.org/10.1016/j.still.2004.03.008
Su M, Han F, Wang M, Ma J, Wang X, Wang Z, Hu S, Li Z (2021) Clay-assisted protection of Enterobacter sp. from Pb (II) stress. Ecotoxicol Environ Saf 208:111704. https://doi.org/10.1016/j.ecoenv.2020.111704
Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH, Sørensen SJ, Karlsson A (2013) 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol 85:612–626. https://doi.org/10.1111/1574-6941.12148
Tan X, Liao H, Shu L, Yao H (2019) Effect of different substrates on soil microbial community structure and the mechanisms of reductive soil disinfestation. Front Microbiol 10:2851
Uroz S, Kelly LC, Turpault M-P, Lepleux C, Frey-Klett P (2015) The mineralosphere concept: mineralogical control of the distribution and function of mineral-associated bacterial communities. Trends Microbiol 23:751–762. https://doi.org/10.1016/j.tim.2015.10.004
Vance ED, Brookes PC, Jenkinson DS (1987) An extraction method for measuring soil microbial biomass C. Soil Biol Biochem 19:703–707. https://doi.org/10.1016/0038-0717(87)90052-6
Vogel C, Babin D, Pronk GJ, Heister K, Smalla K, Kögel-Knabner I (2014) Establishment of macro-aggregates and organic matter turnover by microbial communities in long-term incubated artificial soils. Soil Biol Biochem 79:57–67. https://doi.org/10.1016/j.soilbio.2014.07.012
Wei H, Guenet B, Vicca S, Nunan N, Asard H, AbdElgawad H, Shen W, Janssens IA (2014) High clay content accelerates the decomposition of fresh organic matter in artificial soils. Soil Biol Biochem 77:100–108. https://doi.org/10.1016/j.soilbio.2014.06.006
Weinert N, Meincke R, Gottwald C, Heuer H, Gomes NCM, Schloter M, Berg G, Smalla K (2009) Rhizosphere communities of genetically modified zeaxanthin-accumulating potato plants and their parent cultivar differ less than those of different potato cultivars. Appl Environ Microbiol 75:3859–3865. https://doi.org/10.1128/AEM.00414-09
Whiffen LK, Midgley DJ, McGee PA (2007) Polyphenolic compounds interfere with quantification of protein in soil extracts using the Bradford method. Soil Biol Biochem 39:691–694
Wilkinson JF (1958) The extracellualr polysaccharides of bacteria. Bacteriol Rev 22:46–73
Wingender J, Neu TR, Flemming H-C (1999) What are bacterial extracellular polymeric substances? In: Wingender J, Neu TR, Flemming H-C (eds) Microbial extracellular polymeric substances. Springer, Berlin, pp 1–19
Wright SF, Upadhyaya A (1998) A survey of soils for aggregate stability and glomalin, a glycoprotein produced by hyphae of arbuscular mycorrhizal fungi. Plant Soil 198:97–107. https://doi.org/10.1023/a:1004347701584
Wu J, Joergensen RG, Pommerening B, Chaussod R, Brookes PC (1990) Measurement of soil microbial biomass C by fumigation-extraction-an automated procedure. Soil Biol Biochem 22:1167–1169. https://doi.org/10.1016/0038-0717(90)90046-3
Zethof JH, Bettermann A, Vogel C, Babin D, Cammeraat EL, Solé-Benet A, Lázaro R, Luna L, Nesme J, Woche SK (2020) Prokaryotic community composition and extracellular polymeric substances affect soil microaggregation in carbonate containing semiarid grasslands. Front Environ Sci 8:51
Żur J, Wojcieszyńska D, Guzik U (2016) Metabolic responses of bacterial cells to immobilization. Molecules 21:958
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the Deutsche Forschungsgemeinschaft (DFG):VO2111/1–1 “Microorganisms and the turnover of soil aggregates: the importance of extracellular polymeric substances” and a scholarship funded by the Graduate Academy of the Technische Universität Dresden (Germany) for Folasade Kemi Olagoke. Antje Bettermann was financially supported by the DFG (grant SM 59/18–1 “Extracellular polymeric substances and aggregate stability—how microorganisms affect soil erosion by water”). Doreen Babin was funded by the German Federal Ministry of Education and Research (BMBF) within the framework of the project Di Control (http://dicontrol.igzev.de/en/; research grant 031B0514C).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Olagoke, F.K., Bettermann, A., Nguyen, P.T.B. et al. Importance of substrate quality and clay content on microbial extracellular polymeric substances production and aggregate stability in soils. Biol Fertil Soils 58, 435–457 (2022). https://doi.org/10.1007/s00374-022-01632-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-022-01632-1