Abstract
Purpose
The aim of this study was to develop a model to predict high-genomic-risk prostate cancer (PCa) according to Decipher score, a validated 22 gene prognostic panel. By doing so, one might select the individuals who are likely to benefit from genomic testing and improve pre-op counseling about the need for adjuvant treatments.
Methods
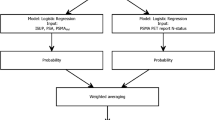
We retrospectively reviewed IRB-approved databases at two institutions. All patients had preoperative magnetic resonance imaging (MRI) and Decipher prostate radical prostatectomy (RP), a validated 22 gene prognostic panel. We used binary logistic regression to estimate high-risk Decipher (Decipher score > 0.60) probability on RP specimen. Area under the curve (AUC) and calibration were used to assess the accuracy of the model in the development and validation cohort. Decision curve analysis (DCA) was performed to assess the clinical benefit of the model.
Results
The development and validation cohort included 622 and 185 patients with 283 (35%) and 80 (43%) of those with high-risk Decipher. The multivariable model included PSA density, biopsy Gleason Grade Group, percentage of positive cores and MRI extracapsular extension. AUC was 0.73 after leave-one-out cross-validation. DCA showed a clinical benefit in a range of probabilities between 15 and 60%. In the external validation cohort, AUC was 0.70 and calibration showed that the model underestimates the actual probability of the outcome.
Conclusions
The proposed model to predict high-risk Decipher score at RP is helpful to improve risk stratification of patients with PCa and to assess the need for additional testing and treatments.


Similar content being viewed by others
Data availability
The data that support the findings of this study are available on request from the corresponding author, [UGF].
References
The L (2018) GLOBOCAN 2018: counting the toll of cancer. Lancet 392(10152):985
Mottet N, van den Bergh RCN, Briers E, Cornford P, De Santis M, Fanti S et al (2022) EAU - ESTRO - ESUR - SIOG Guidelines on Prostate Cancer 2022. European Association of Urology Guidelines 2022 Edition. presented at the EAU Annual Congress Amsterdam 2022. Arnhem, The Netherlands: European Association of Urology Guidelines Office
NCCN (2021) Prostate Cancer (Version 2.2022). https://www.nccn.org/professionals/physician_gls/pdf/prostate_blocks.pdf
Heidegger I, Klocker H, Pichler R, Pircher A, Prokop W, Steiner E et al (2017) ProPSA and the Prostate Health Index as predictive markers for aggressiveness in low-risk prostate cancer-results from an international multicenter study. Prostate Cancer Prostatic Dis 20(3):271–275
de Cobelli O, Terracciano D, Tagliabue E, Raimondi S, Bottero D, Cioffi A et al (2015) Predicting pathological features at radical prostatectomy in patients with prostate cancer eligible for active surveillance by multiparametric magnetic resonance imaging. PLoS One 10(10):e0139696
Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA et al (2016) The 2014 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma: definition of grading patterns and proposal for a new grading system. Am J Surg Pathol 40(2):244–252
Den RB, Yousefi K, Trabulsi EJ, Abdollah F, Choeurng V, Feng FY et al (2015) Genomic classifier identifies men with adverse pathology after radical prostatectomy who benefit from adjuvant radiation therapy. J Clin Oncol 33(8):944–951
Klein EA, Yousefi K, Haddad Z, Choeurng V, Buerki C, Stephenson AJ et al (2015) A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur Urol 67(4):778–786
Van den Broeck T, Moris L, Gevaert T, Tosco L, Smeets E, Fishbane N et al (2019) Validation of the Decipher test for predicting distant metastatic recurrence in men with high-risk nonmetastatic prostate cancer 10 years after surgery. Eur Urol Oncol 2(5):589–596
Spratt DE, Zhang J, Santiago-Jimenez M, Dess RT, Davis JW, Den RB et al (2018) Development and validation of a novel integrated clinical-genomic risk group classification for localized prostate cancer. J Clin Oncol 36(6):581–590
Rosenkrantz AB, Ginocchio LA, Cornfeld D, Froemming AT, Gupta RT, Turkbey B et al (2016) Interobserver reproducibility of the PI-RADS version 2 lexicon: a multicenter study of six experienced prostate radiologists. Radiology 280(3):793–804
Falagario U, Ratnani P, Lantz A, Jambor I, Dovey Z, Verma A et al (2020) Staging accuracy of multiparametric MRI in Caucasian and African American patients undergoing radical prostatectomy. J Urol. https://doi.org/10.1097/JU0000000000000774
Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK et al (2017) The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin 67(2):93–99
Den RB, Santiago-Jimenez M, Alter J, Schliekelman M, Wagner JR, Renzulli Ii JF et al (2016) Decipher correlation patterns post prostatectomy: initial experience from 2342 prospective patients. Prostate Cancer Prostatic Dis 19(4):374–379
Assel M, Sjoberg D, Elders A, Wang X, Huo D, Botchway A et al (2019) Guidelines for reporting of statistics for clinical research in urology. J Urol 201(3):595–604
Reese AC, Feng Z, Landis P, Trock BJ, Epstein JI, Carter HB (2015) Predictors of adverse pathology in men undergoing radical prostatectomy following initial active surveillance. Urology 86(5):991–995
Park JW, Koh DH, Jang WS, Cho KS, Ham WS, Rha KH et al (2018) Predictors of adverse pathologic features after radical prostatectomy in low-risk prostate cancer. BMC Cancer 18(1):545
Audenet F, Vertosick EA, Fine SW, Sjoberg DD, Vickers AJ, Reuter VE et al (2018) Biopsy core features are poor predictors of adverse pathology in men with grade group 1 prostate cancer. J Urol 199(4):961–968
Morselli S, Sebastianelli A, Campi R, Liaci A, Gabellini L, Tasso G et al (2019) Adverse pathology after radical prostatectomy: the prognostic role of cumulative cancer length >6-mm threshold in prostate cancer-positive biopsies. Prostate Int 7(4):143–149
Falagario UG, Jambor I, Lantz A, Ettala O, Stabile A, Taimen P et al (2020) Combined use of prostate-specific antigen density and magnetic resonance imaging for prostate biopsy decision planning: a retrospective multi-institutional study using the prostate magnetic resonance imaging outcome database (PROMOD). Eur Urol Oncol 4(6):971–979
Lantz A, Falagario UG, Ratnani P, Jambor I, Dovey Z, Martini A et al (2022) Expanding active surveillance inclusion criteria: a novel nomogram including preoperative clinical parameters and magnetic resonance imaging findings. Eur Urol Oncol 5(2): 187–194
Cumarasamy S, Martini A, Falagario UG, Gul Z, Beksac AT, Jayaratna I et al (2020) Development of a model to predict prostate cancer at the apex (PCAP model) in patients undergoing robot-assisted radical prostatectomy. World J Urol 38(4):813–819
Jambor I, Falagario U, Ratnani P, Perez IM, Demir K, Merisaari H et al (2020) Prediction of biochemical recurrence in prostate cancer patients who underwent prostatectomy using routine clinical prostate multiparametric MRI and Decipher genomic score. J Magn Reson Imaging 51(4):1075–1085
Li L, Shiradkar R, Leo P, Algohary A, Fu P, Tirumani SH et al (2021) A novel imaging based Nomogram for predicting post-surgical biochemical recurrence and adverse pathology of prostate cancer from pre-operative bi-parametric MRI. EBioMedicine 63:103163
Beksac AT, Cumarasamy S, Falagario U, Xu P, Takhar M, Alshalalfa M et al (2018) Multiparametric magnetic resonance imaging features identify aggressive prostate cancer at the phenotypic and transcriptomic level. J Urol 200(6):1241–1249
Jairath NK, Dal Pra A, Vince R Jr, Dess RT, Jackson WC, Tosoian JJ et al (2021) A systematic review of the evidence for the Decipher genomic classifier in prostate cancer. Eur Urol 79(3):374–383
Feng FY, Huang HC, Spratt DE, Zhao SG, Sandler HM, Simko JP et al (2021) Validation of a 22-gene genomic classifier in patients with recurrent prostate cancer: an ancillary study of the NRG/RTOG 9601 randomized clinical trial. JAMA Oncol 7(4):544–552
Dal Pra A, Ghadjar P, Hayoz S, Spratt DE, Liu VYT, Todorovic T et al (2021) Validation of the Decipher genomic classifier (GC) in SAKK 09/10: a phase III randomized trial of dose-escalated salvage radiotherapy (SRT) after radical prostatectomy (RP). J Clin Oncol 39(15_suppl):5010
Nguyen PL, Huang HC, Davicioni E, Sandler HM, Shipley WU, Efstathiou JA et al (2021) Validation of a 22-gene genomic classifier in the NRG oncology/RTOG 9202, 9413 and 9902 phase III randomized trials: a biopsy-based individual patient meta-analysis in high-risk prostate cancer. Int J Radiat Oncol Biol Phys 111(3):S50
Acknowledgements
This research was supported by a grant of the European Urological Scholarship Programme awarded to UGF.
Funding
None.
Author information
Authors and Affiliations
Contributions
UGF, AK, MS and AL conceived and designed the study; PR, SP, KH, VW, DC and AE acquired the data; UGF, AM and DG analyzed and interpreted the data; UGF, MS, AL, AM and IJ drafted the manuscript; AT, GC, LC, NK, PW, MWK and EAK critically revised the manuscript for important intellectual content; DL provided administrative, technical, or material support; AT, GC, DL, PW, MWK and EAK took part in supervision; and KH carried out the pathologic examination of the slides.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest related to the present study.
Ethical approval and consent to participate
The present study was approved by Mount Sinai Hospital ethical committee (IRB-19-01597) with a waiver on consent to participate.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Figure 1.
Calibration plot of observed versus predicted probability of biologically significant prostate cancer (high-risk Decipher) according to the risk calculator in the development and validation cohort. Calibration plot of the development cohort is shown for the predicted probability of the model after Logistic regression (LR) and after leave-one-out cross-validation (LOOCV). (TIFF 10073 KB)
Supplementary Figure 2.
Decision curve analysis (DCA) demonstrating the net benefit associated with the use of the risk calculator-derived probability for the prediction of high-risk Decipher score in the development cohort. DCA is a statistical method evaluating the clinical benefit of a model when used for clinical decision making. In our case, the clinical decision is to perform or not a genomic test. Two reference strategies are defined: i. performing Decipher test in all and ii. performing Decipher test in no one. The third line refers to perform Decipher test using the model predicted probability. The range of probability where this line is above the reference lines defines the upper and the lower limit of probability where the model has a clinical benefit. (TIF 13928 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Falagario, U.G., Chakravarty, D., Martini, A. et al. When to order genomic tests: development and external validation of a model to predict high-risk prostate cancer at the genotypic level. World J Urol 41, 85–92 (2023). https://doi.org/10.1007/s00345-022-04240-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00345-022-04240-8