Abstract
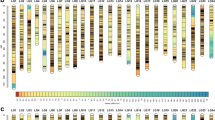
Ancherythroculter nigrocauda is a fish endemic to the upper areas of the Changjiang (Yangtze) River in China. Quantitative trait locus (QTL) mapping is a powerful tool to identify potential genes affecting traits of economic importance in domestic animals. In this study, a high-density genetic map was constructed with 5 901 single nucleotide polymorphism (SNP) makers by sequencing 92 individual fish from a F1 family using the specific-locus amplified fragment sequencing approach. Initially, 48 QTLs for total length, body length, body height, and body weight were identified according to the high density of the genetic map with 24 LGs, a total length of 3 839.4 cM, and marker spacing of about 0.82 cM. These QTLs explained 27.1%–49.9% of phenotypic variance. The results of this study suggest that major QTLs are responsible for the growth of A. nigrocauda, and these are potentially useful in comparative genomics research, genome assembly, and marker-assisted breeding programs for this species.
Similar content being viewed by others
Data Availability Statement
All data generated and/or analyzed during the study are available from the corresponding author on reasonable request.
References
Andrews K R, Good J M, Miller M R, Luikart G, Hohenlohe P A. 2016. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat. Rev. Genet., 17(2): 81–92.
Bai Z Y, Han X K, Liu X J, Li Q Q, Li J L. 2016. Construction of a high-density genetic map and QTL mapping for pearl quality-related traits in Hyriopsis cumingii. Sci. Rep., 6: 32608.
Baird N A, Etter P D, Atwood T S, Currey M C, Shiver A L, Lewis Z A, Selker E U, Cresko W A, Johnson E A. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoSOne, 3(10): e3376.
Chakravarti A, Lasher L K, Reefer J E. 1991. A maximum likelihood method for estimating genome length using genetic linkage data. Genetics, 128(1): 175–182.
Davey J W, Hohenlohe P A, Etter P D, Boone J Q, Catchen J M, Blaxter M L. 2011. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet., 12(7): 499–510.
Dib C, Fauré S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Kazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J. 1996. A comprehensive genetic map of the human genome based on 5264 microsatellites. Nature, 380(6570): 152–154.
Faris J D, Laddomada B, Gill B S. 1998. Molecular mapping of segregation distortion loci in Aegilops tauschii. Genetics, 149(1): 319–327.
Feng X, Yu X M, Fu B D, Wang X H, Liu HY, Pang M X, Tong J G. 2018. A high-resolution genetic linkage map and QTL fine mapping for growth-related traits and sex in the Yangtze River common carp (Cyprinus carpio haematopterus). BMC Genomics, 19: 230.
Fishman L, Kelly A J, Morgan E, Willis J H. 2001. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to hetero specific interactions. Genetics, 159(4): 1 701–1 716.
Fu B D, Liu H Y, Yu X M, Tong J G. 2016. A high-density genetic map and growth related QTL mapping in bighead carp (Hypophthalmichthys nobilis). Sci. Rep., 6: 28 679.
Gjedrem T. 2000. Genetic improvement of cold-water fish species. Aquac. Res., 31(1): 25–33.
Guo J Q, Li C J, Teng T, Shen F F, Chen Y N, Wang Y F, Pan C L, Ling Q F. 2018. Construction of the first high-density genetic linkage map of pikeperch (Sander lucioperca) using specific length amplified fragment (SLAF) sequencing and QTL analysis of growth-related traits. Aquaculture, 497: 299–305.
Huang X H, Zhao Y, Wei X H, Li C Y, Wang A H, Zhao Q, Li W J, Guo Y L, Deng L W, Zhu C R, Fan D L, Lu Y Q, Weng Q J, Liu K Y, Zhou T Y, Jing Y F, Si L Z, Dong G J, Huang T, Lu T T, Feng Q, Qian Q, Li J Y, Han B. 2011. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet., 44(1): 32–39.
Ihara N, Takasuga A, Mizoshita K, Takeda H, Sugimoto M, Mizoguchi Y, Hirano T, Itoh T, Watanabe T, Reed K M, Snelling W M, Kappes S M, Beattie C W, Bennett G L, Sugimoto Y. 2004. A comprehensive genetic map of the cattle genome based on 3802 microsatellites. Genome Res., 14(10A): 1 987–1 998.
Jansen J, de Jong A G, van Ooijen J W. 2001. Constructing dense genetic linkage maps. Theor. Appl. Genet., 102: 1 113–1 122.
Kosambi D D. 1943. The estimation of map distances from recombination values. Ann. Eugen., 12(1): 172–175.
Lallias D, Beaumont A R, Haley C S, Boudry P, Heurtebise S, Lapègue S. 2007. A first-generation genetic linkage map of the European flat oyster Ostreaedulis (L.) based on AFLP and microsatellite markers. Anim. Genet., 38(6): 560–568.
Li L, Xiang J H, Liu X, Zhang Y, Dong B, Zhang X J. 2005. Construction of AFLP-based genetic linkage map for Zhikong scallop, Chlamys farreri Jones et Preston and mapping of sex-linked markers. Aquaculture, 245(1–4): 63–73.
Li R Q, Li Y R, Kristiansen K, Wang J. 2008. SOAP: short oligonucleotide alignment program. Bioinformatics, 24(5): 713–714.
Liu C C, Gao X, Wang H S, Liu H Z, Cao W X, Danley P D. 2013. Reproductive characteristics of Ancherythroculter nigrocauda, an endemic fish in the upper Yangtze River, China. Fisheries Sci., 79(5): 799–806.
Liu D Y, Ma C X, Hong W G, Huang L, Liu M, Liu H, Zeng H P, Deng D J, Xin H G, Song J, Xu C H, Sun X W, Hou X L, Wang X W, Zheng H K. 2014. Construction and analysis of high-density linkage map using high-throughput sequencing data. PLoSOne, 9(6): e98855.
Liu H Y, Fu B D, Pang M X, Feng X, Yu X M, Tong J G. 2017. A high-density genetic linkage map and QTL fine mapping for body weight in crucian carp (Carassius auratus) using 2b-RAD sequencing. G3, 7(8): 2 473–2 487.
Liu J K, Cao W X. 1992. Fish resources of the Yangtze River basin and the tactics for their conservation. Resour. Environ. Yangtze Basin, 1(1): 17–23. (in Chinese with English abstract)
Niu D H, Du Y C, Wang Z, Xie S M, Nguyen H, Dong Z G, Shen H D, Li J L. 2017. Construction of the first high-density genetic linkage map and analysis of quantitative trait loci for growth-related traits in Sinonovacula constricta. Mar. Biotechnol., 19(5): 488–496.
Peng W Z, Xu J, Zhang Y, Feng J X, Dong C J, Jiang L K, Feng J Y, Chen B H, Gong Y W, Chen L, Xu P. 2016. An ultrahigh density linkage map and QTL mapping for sex and growth-related traits of common carp (Cyprinus carpio). Sci. Rep., 6: 26 693.
Peterson B K, Weber J N, Kay E H, Fisher H S, Hoekstra H E. 2012. Double Digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoSOne, 7(5): e37135.
Qiu G F, Xiong L W, Han Z K, Liu Z Q, Feng J B, Wu X G, Yan Y L, Shen H, Huang L, Chen L. 2017. A second generation SNP and SSR integrated linkage map and QTL mapping for the Chinese mitten crab Eriocheir sinensis. Sci. Rep., 7: 39 826.
Sambrook J, Russell D W. 2001. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor Laboratory Press, New York. p.463–446.
Shao C W, Niu Y C, Rastas P, Liu Y, Xie Z Y, Li H D, Wang L, Jiang Y, Tai S S, Tian Y S, Sakamoto T, Chen S L. 2015. Genome-wide SNP identification for the construction of a high-resolution genetic map of Japanese flounder (Paralichthys olivaceus): applications to QTL mapping of Vibrio anguillarum disease resistance and comparative genomic analysis. DNA Res., 22(2): 161–170.
Singer A, Perlman H, Yan Y L, Walker C, Corley-Smith G, Brandhorst B, Postlethwait J. 2002. Sex-specific recombination rates in zebrafish (Danio rerio). Genetics, 160(2): 649–657.
Sun C F, Niu Y C, Ye X, Dong J J, Hu W S, Zeng Q K, Chen Z H, Tian Y Y, Zhang J, Lu M X. 2017. Construction of a high-density linkage map and mapping of sex determination and growth-related loci in the mandarin fish (Siniperca chuatsi). BMC Genomics, 18: 446.
Sun X W, Liu D Y, Zhang X F, Li W B, Liu H, Hong W G, Jiang C B, Guan N, Ma C X, Zeng H P, Xu C H, Song J, Huang L, Wang C M, Shi J J, Wang R, Zheng X H, Lu C Y, Wang X W, Zheng H K. 2013. SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoSOne, 8(3): e58700.
Sun Y H, Li Q, Wang G Y, Zhu D M, Chen J, Li P. 2015. Development of transcript-associated microsatellite markers in Ancherythoculter nigrocauda and cross-amplification in Culter alburnus. Genet. Mol. Res., 14(4): 14 286–14 290.
Sun Y H, Li Q, Wei H J, Wang G Y, Chen J, Li P. 2018. Single nucleotide polymorphism identification in growth-related genes from the transcriptome of the fish Ancherythroculter nigrocauda. Conserv. Genet. Resour., 10(2): 153–155.
Sun Y H, Wang G Y, Zhu D M, Chen J, Li P, Li Q. 2014. Development of polymorphic microsatellite loci isolated from the Ancherythoculter nigrocauda. Conserv. Genet. Resour., 6(4): 919–923.
Tong J G, Sun X W. 2015. Genetic and genomic analyses for economically important traits and their applications in molecular breeding of cultured fish. Sci. China Life Sci., 58(2): 178–186.
Van Ooijen J. 2011. Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet. Res., 93(5): 343–349.
Van Os H, Stam P, Visser R G, van Eck H J. 2005. SMOOTH: a statistical method for successful removal of genotyping errors from high-density genetic linkage data. Theor. Appl. Genet., 112(1): 187–194.
Voorrips R E. 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered., 93(1): 77–78.
Wang L, Wan Z Y, Bai B, Huang S Q, Chua E, Lee M, Pang H Y, Wen Y F, Liu P, Liu F, Sun F, Lin G, Ye B Q, Yue G H. 2015. Construction of a high-density linkage map and fine mapping of QTL for growth in Asian seabass. Sci. Rep., 5: 16 358.
Wang S, Meyer E, McKay J K, Matz M V. 2012. 2b-RAD: a simple and flexible method for genome-wide genotyping. Nat. Methods, 9(8): 808–810.
Xia J H, Lin G, He X P, Yunping B, Liu P, Liu F, Sun F, Tu R J, Yue G H. 2014. Mapping quantitative trait loci for omega-3 fatty acids in Asian seabass. Mar. Biotechnol., 16(1): 1–9.
Xu S Z. 2008. Quantitative trait locus mapping can benefit from segregation distortion. Genetics, 180(4): 2 201–2 208.
Young W P, Wheeler P A, Coryell V H, Keim P, Thorgaard G H. 1998. A detailed linkage map of rainbow trout produced using doubled haploids. Genetics, 148(2): 839–850.
Yu Y, Zhang X J, Yuan J B, Li F H, Chen X H, Zhao Y Z, Huang L, Zheng H K, Xiang J H. 2015. Genome survey and high-density genetic map construction provide genomic and genetic resources for the Pacific White Shrimp Litopenaeus vannamei. Sci. Rep., 5: 15 612.
Yu Z N, Guo X M. 2003. Genetic linkage map of the eastern oyster Crassostrea virginica Gmelin. Biol. Bull., 204(3): 327–338.
Yue G H. 2014. Recent advances of genome mapping and marker-assisted selection in aquaculture. Fish and Fisheries, 15(3): 376–396.
Zhang J, Zhang Q X, Cheng T R, Yang W R, Pan H T, Zhong J J, Huang L, Liu E Z. 2015. High-density genetic map construction and identification of a locus controlling weeping trait in an ornamental woody plant (Prunusmume Sieb. et Zucc). DNA Res., 22(3): 183–191.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supported by the Technical Innovation Project of Hubei Province (No. 2018ABA105) and the Enterprise Technology Innovation Project of Wuhan (No. 39 of 2019 WuKe)
Rights and permissions
About this article
Cite this article
Sun, Y., Li, P., Wang, G. et al. Construction of the first high-density genetic map for growth related QTL analysis in Ancherythroculter nigrocauda. J. Ocean. Limnol. 39, 1118–1130 (2021). https://doi.org/10.1007/s00343-020-9290-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00343-020-9290-7