Abstract
Microorganisms have been reported to induce settlement in various marine invertebrate larvae but their specificity of inductive capacities for the settlement of coral larvae remains poorly understood. In this study, we isolated 56 microbial strains from the crustose coralline alga (CCA) Hydrolithon reinboldii using five different media either with or without additional antibiotics and/or spiked CCA extract. We tested the isolates for their potential to induce settlement behavior in larvae of the brooding scleractinian coral Leptastrea purpurea. From these 56 CCA-associated microbial strains, we identified six bacterial classes and 18 families. The culturable bacterial community associated with H. reinboldii was dominated by Gammaproteobacteria, Actinobacteria, and Alphaproteobacteria while the Illumina MiSeq analysis showed that the culture-independent bacterial community was dominated by Gammaproteobacteria, Alphaproteobacteria, and Flavobacteria. Furthermore, we found no correlation between inductive settlement capacities and phylogenetic relationships. Instead, settlement behavior of L. purpurea larvae was induced by specific isolated species. Strains #1792 (Pseudovibrio denitrificans), #1678 (Acinetobacter pittii), #1633 (Pseudoalteromonas phenolica), #1772 (Marine bacterium LMG1), #1721 (Microbulbifer variabilis), and #1783 (Pseudoalteromonas rubra) induced settlement behavior in coral larvae at mostly high and significant levels (≥ 40%) but the remaining isolates strongly varied in their activity. Multispecies biofilms consisting of four strains (#1792, #1678, #1633, and #1721) were observed to synergistically increase settlement behavior levels (> 90%); however, the addition of #1772 to the multispecies biofilms negatively affected coral larvae and resulted in a total loss of inducing activity. The findings provide new insights into the role of bacteria in the settlement process of scleractinian corals and may help to identify the true nature of bacteria-derived morphogenic cues.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coral reefs are among the world’s most diverse ecosystems. They serve as nursery grounds and feeding areas for many reef-dependent animal species and affect humankind by having major impacts on economy and even politics (Pauly et al. 2002; Bellwood et al. 2004; Barbier et al. 2011). Unfortunately, coral reefs are also highly threatened ecosystems mainly due to climate change leading to severe bleaching events on a global scale (Hoegh-Guldberg et al. 2007; Heron et al. 2016; Ainsworth et al. 2016). Because threatened coral reef systems depend on the recruitment of new individuals for recovery (Edmunds 2004; Negri et al. 2007), a better understanding of the recovery and population dynamics of stony corals is necessary. The recruitment process can be divided into (i) the development of competent larvae in the water column (spawning corals) or within the corals itself (brooding corals), (ii) the settlement process (i.e., searching, attachment, and metamorphosis) onto suitable substrates, and (iii) the survival of juvenile corals (Ritson-Williams et al. 2009). Since the survival rate of juvenile corals is likely influenced by the type of substrate chosen for settlement (Harrington et al. 2004; Ritson-Williams et al. 2010), finding a suitable settlement ground may be the most critical step within the recruitment process. It is widely accepted that the transition from planktonic to benthic life stages of invertebrate larvae depends on settlement cues that signal habitat suitability. These cues can be of biological, physical, and/or chemical nature, bearing information about the settlement habitat, or might come in the form of environmental parameters. Many studies have shown that coral larvae settle in response to either live crustose coralline algae (CCA) (Morse et al. 1988, 1991; Price 2010; Ritson-Williams et al. 2014) or organic crude extracts of the CCA (Heyward and Negri 1999; Harrington et al. 2004; Kitamura et al. 2009; Tebben et al. 2015). For example, Tebben and colleagues (2015) isolated two classes of CCA cell wall-associated compounds, glycoglycerolipids and polysaccharides, which induced settlement (attachment and metamorphosis) in Acropora millepora larvae at equivalent concentrations present in live CCA. However, ubiquitous microbial biofilms, covering the surface of marine hard substrata such as living CCA, have received attention too. Many studies provided evidence that microbial biofilms are potent inducers of settlement within a broad variety of invertebrate larvae, including echinoderms, polychaetes, molluscs, barnacles, bryozoans, and cnidaria (Johnson et al. 1991; Pawlik 1992 and referenced therein; Dobretsov et al. 2006; Thiyagarajan et al. 2006; Hadfield 2011 and referenced therein). Specifically, larvae of scleractinians have been found to selectively settle on natural biofilms, containing, for example, a variety of epiphytic bacteria and microalgae but also on monospecific bacterial biofilms isolated from CCA (Negri et al. 2001; Webster et al. 2004; Erwin et al. 2008). Tetrabromopyrrole, a secondary metabolite isolated from a CCA-associated Pseudoalteromonas strain, induced metamorphosis without prior attachment in larvae of the spawning coral A. millepora (Tebben et al. 2011). Further studies revealed that this compound induces settlement (attachment and metamorphosis) in larvae of the brooding coral Porites astreoides as well as in larvae of the broadcast-spawning corals Orbicella franski and Acropora palmata (Sneed et al. 2014). These discoveries gave first insights into the broad range of morphogenic cues and demonstrate the role that microorganisms may play in the coral recruitment process. However, some reported biofilms did induce metamorphosis without prior attachment and thus resulted in unsuccessful settlement (Negri et al. 2001; Tebben et al. 2011). In addition, their effect on coral settlement was often tested using natural biofilms with largely unknown bacterial compositions that had developed on glass slides in the water column (Webster et al. 2004; Erwin et al. 2008). Thus, only a few studies investigated the diversity of culturable bacteria associated with CCA and have proven the ability of monospecific biofilms to induce complete settlement in scleractinian coral larvae (Tran and Hadfield 2011; Sneed et al. 2014). In continuation of our research interest into the recruitment and survival of reef-building stony corals (Kitamura et al. 2009; Moeller et al. 2017, 2019; Nietzer et al. 2018), we aimed to isolate bacterial strains from the biofilm associated with the CCA Hydrolithon reinboldii and systematically test each bacterial isolate, as well as combinations of selected isolates, for its ability to induce settlement behavior (attachment and/or metamorphosis) in larvae of the scleractinian brooding coral Leptastrea purpurea.
Material and methods
Collection of crustose coralline algae (CCA) and Leptastrea purpurea corals
All CCA and Leptastrea purpurea corals were collected snorkeling from a depth between one and two meters between April and July 2010 in Luminao, Guam (13°27′53″ N, 144°38′54″ E). CCA pieces of Hydrolithon reinboldii, which had completely overgrown dead coral rubble, were collected by hand. CCA were identified by comparing collected specimen with voucher material at the UOG (University of Guam) Marine Laboratory. Dr. Mark Littler, a known CCA taxonomist, had previously identified and deposited CCA vouchers at the UOG Marine Laboratory collection. Identification based on the voucher material was conducted by Dr. Peter Schupp and additional vouchers were deposited to the Marine Laboratory collection. Furthermore, 80 colonies of L. purpurea were cautiously detached from substrate with hammer and chisel. After sampling, both CCA and corals were transported to the Marine Laboratory facility at the University of Guam and placed in 500 l basins with a steady flow-through of sea water from Pago Bay (13°25′36″ N, 144°47′56″ E) at ambient temperature (28–29 °C). After two days of acclimation, CCA and coral colonies were transferred to a 3500 l flow-through tank for larval collection. The CCA were kept in the tank for one week and L. purpurea corals for a duration of four months until replanting both to the Luminao Reef.
Isolation and identification of bacteria
Media for the isolation of bacteria were inoculated the same day that the CCA H. reinboldii were collected. For this purpose, agar plates of four nutrient-rich and one minimal media (M1, M5, AC, 1:1 MB, and M8, respectively) were used for bacterial isolation (see list of media recipes in online resource 1 for details). Furthermore, additional agar plates of both M1 and 1:1 MB media were supplemented with rifampicin (5 µg ml−1) and Gentamicin (2 µg ml−1), two broad-spectrum antibiotics. Additionally, further agar plates of 1:1 MB, AC, and M8 media were supplemented with CCA extract and the two broad-spectrum antibiotics. The extract was obtained by extracting 3.2 kg of CCA pieces which were completely overgrown by H. reinboldii with 70% EtOH for 5 h. Four iterations of this process yielded 26.5 g crude extract. After autoclave sterilization, the media were supplemented with CCA extract, using two parts by weight of extract dissolved in one part by weight of EtOH. The weight of the CCA extract added to 1 l medium equates to the weight of CCA extract extracted from 1 kg CCA. Therefore, 8.28 g CCA extract and 4.14 g EtOH were added to 1 l of the respective medium. Table 1 shows all used media for the isolation of the CCA-associated bacteria. In general, four techniques were used for bacterial isolation on all media types shown in Table 1: (i) the surface of the CCA was scraped off using a sterile scalpel. The algae biomass had been diluted with autoclaved, filtered seawater (80 µm, Whatman, GE Healthcare, Chalfont St Giles, UK) and the following dilution series (1:1, 1:10, 1:100, and 1:1000) of the CCA suspension was spread on agar plates using 100 µl, respectively. (ii) A further dilution series (similar as above) of CCA suspension was heated to 55 °C for 10 min in order to isolate spore-forming bacteria and spread onto agar plates. (iii) Sterile cotton swabs wiped the surface of the CCA for 15 s. Afterward, the swabs were used for dilution streaks on agar plates. (iv) Small pieces of dead coral rubble (approx. 4 cm) overgrown with CCA were touched to the agar surface three times and then left on the plates. All four isolation techniques were applied in triplicate. For initial isolations of bacteria from CCA, cycloheximide (100 µg ml−1) was added to each medium after autoclave sterilization. The inoculated Petri dishes were stored for seven days at 27 °C. All plates were checked for different morphotypes as well as for the amount of colony forming units (CFUs) on a daily basis. After the first week, plates were continuously checked for new colonies every three days. This monitoring was carried out for one month. All different morphotypes detected during evaluations were isolated by streaking on fresh agar plates of the corresponding media to obtain axenic cultures. The bacterial isolates were identified by 16S rRNA gene Sanger sequencing. Therefore, DNA of most strains was extracted using the freeze–thaw method. Cell material from agar plates was sampled and mixed with 40 µl nuclease-free water. After a 5-s vortex step, the suspension was incubated at − 20 °C for 30 min. DNA of an actinomycete was not accessible with this method and was therefore extracted using the Qiagen DNeasy Extraction Kit (Qiagen, Venlo, The Netherlands). Molecular amplification of the 16 s rRNA region was performed using the forward primer 27f and the reverse primer 1492r (Jiang et al. 2006). Per sample, the PCR mixture consisted of 35 µl filter-sterilized Omnipur H2O, 5 µl 10 × buffer, 2 µl 27f primer, 2 µl 1492r primer, 4 µl dNTP’s, 0.5 µl Taq-polymerase, and 1.5 µl of extracted DNA. The program and settings of the amplification contained the following steps: initial denaturation at 95 °C for 5 min, 30 cycles of denaturation at 95 °C for 60 s, annealing at 55 °C for 60 s, elongation at 72 °C for 1 min 30 s, a final elongation at 72 °C for 10 min, and cooling at 4 °C. Sanger sequencing was performed by MacroGen (Seoul, South Korea) using 27f as the sequencing primer. Reads were trimmed with DNA Baser version 3.5.2.4 until there were 99% good bases (quality value < 21) in each 20-base window. Subsequently, the reads were compared by BLASTn (Altschul et al. 1990) to the 16S rRNA genes in the SILVA database of type strains (Quast et al. 2013).
DNA extraction and MiSeq microbiome analyses
For the investigation of the non-culturable CCA-associated bacterial communities, loose CCA fragments (3–5 cm in length and ca. 1 cm in diameter) were collected in sterile falcon tubes from sand and coral rubble patches at Luminao Reef (n = 3) to assess the overall CCA microbiome. In addition, the surface of the CCA fragments was wiped underwater with sterile cotton swabs (n = 3) to determine the surface-associated microbiome. Surface sediments were collected underwater by scraping the top 1 cm into sterile falcon tubes (n = 3) for comparison with the CCA-associated microbiome. All samples were preserved in 100% EtOH in the field, transported in a cooling box to the UOG laboratory and there stored at − 20 °C until DNA isolation was carried out using the Qiagen DNeasy PowerSoil Kit. CCA fragments were crushed in a sterile mortar previous to processing. Prior to DNA isolation, cell material was centrifuged, and the supernatant was removed. For each sample, a barcoded 16S rRNA gene PCR was performed with primers amplifying a 292 bp fragment in the V4 region as previously described (Kozich et al. 2013). The composite forward primer consisted of the Illumina 5′ adapter, a 8-nt barcode, a 10-nt pad sequence, a 2-nt linker and the 515F-Y 16S rRNA gene-specific primer (Parada et al. 2016), whereas the composite reverse primer consisted of the Illumina 3′ adapter, a 8-nt barcode, a 10-nt pad sequence, a 2-nt linker, and the 806rB 16S rRNA gene-specific primer (Apprill et al. 2015). PCR amplifications were performed in a reaction volume of 25 µl containing 5 µl Green GoTaq® reaction buffer (Promega, Madison, USA), 0.5 µl 10 mM dNTPs (Promega), 0.5 µl 10 µM forward primer, 0.5 µl 10 µM reverse primer, 0.15 µl GoTaq® DNA polymerase (5U µl−1, Promega) and 1 µl template DNA (0.1–10 ng µl−1). The PCR program consisted of: initial denaturation of 2 min at 95 °C; 30 cycles of denaturation at 95 °C for 20 s, annealing at 55 °C for 15 s, and extension at 72 °C for 2 min; and final extension at 72 °C for 10 min. Samples were amplified in triplicate, after which the reaction volumes were pooled and 5 µl combined solution was run on a 1% agarose gel to assess amplification success. Subsequently, the PCR products were purified using the QIAquick PCR Purification Kit (Qiagen). DNA was eluted from the spin column with 10 µl distilled DNase/RNase-free water (Invitrogen, Waltham, USA). The DNA concentration of the elute was measured with the Nanodrop 2000c (Thermo Fisher Scientific, Waltham, USA). An equimolar mixture of PCR products from unique samples, i.e., a sample library, was prepared and run on a 1% agarose gel. The gel band at ~ 292 bp was extracted and purified using the ZymocleanTM Gel DNA Recovery Kit (Zymo Research, Irvine, USA). Elution was done with distilled DNase/RNase-free water. The sample mixture was reduced in volume by vacuum drying and subsequently sent for Illumina paired end MiSeq sequencing (2 × 250 bp) at GATC Biotech. A negative control sample, which was obtained via a PCR reaction without template DNA, was also included in the sample library because weak amplification was regularly detected in PCRs without template. The 16S rRNA gene amplicon sequences were deposited in the ENA SRA database under accession number PRJEB31052 (see also Table 2).
Processing of amplicon data
The 16S rRNA gene reads were processed with the MiSeq standard operating procedure of the open source software package mothur (Kozich et al. 2013) (https://www.mothur.org/wiki/MiSeq_SOP, accessed November 30, 2017). In brief, reads were quality-filtered, assembled into contigs, filtered for chimeras with the VSEARCH algorithm (Rognes et al. 2016), and clustered into operational taxonomic units (OTUs) based on a 97% identity threshold. The OTUs were annotated with the Ribosomal Database Project Classifier (Wang et al. 2007) using the SILVA SSU NR 128 database as a reference (Quast et al. 2013). OTUs that were detected at a higher relative abundance in the negative control sample than in our biological samples were removed from the OTU table because they were assumed to represent contamination. R version 3.4.3 (R development Core Team) was used to execute R packages. R package “phyloseq” version 1.22.3 was used to rarefy samples to even depth. Hereafter, microbiome data from the Illumina MiSeq analysis are given in mean + standard error (SE).
Settlement behavior assays
Colonies were placed onto trays made of egg crate for better handling and collection of larvae was conducted as described in Nietzer and colleagues (2018). Monospecific biofilms of the isolated bacteria were grown on sterile coverslips and tested for coral larvae settlement behavior. Therefore, all isolates were grown in liquid 1:1 MB media (without agar) for 24–48 h. Three sterile coverslips were placed in one sterile Petri dish, covered with the respective liquid bacterial culture and incubated for 24 h at 22 °C to ensure biofilm formation. After the 24 h incubation period, each cover slip was placed with the overgrown side facing upward into a sterile Petri dish filled with 10 ml filtered and sterilized seawater. In most cases, biofilms grew evenly and were clearly visible. However, in some cases where strains grew patchy and coagulative, bacterial cell agglomerates attached to the top of the coverslips were placed together with the coverslips into the Petri dishes. To generate multispecies biofilms, selected strains were cultured in liquid media as described above and following 24–48 h incubation, 10 ml from each culture were mixed together. Finally, the bacterial mixture was poured onto three coverslips. Further steps to generate multispecies biofilms proceeded as described above for the generation of monospecific biofilms. As positive controls, four pieces of fresh H. reinboldii (ca. 3 × 3 × 2 mm) on top of a sterile coverslip were added to a sterile Petri dish filled with 10 ml FSW as positive control. The negative controls were prepared in the same way without CCA pieces. Each bacterial biofilm, including the multispecies biofilms, was tested in triplicate (n = 3), with each replicate containing five randomly selected L. purpurea larvae mixed from all 80 coral colonies. All larvae were between one and three days old and therefore well within the timeframe where no difference in settlement behavior should be observed (see Nietzer et al. 2018). Replicates of the positive and negative controls were treated equally; however, there were not enough larvae to perform all experiments in one day or to perform controls for each individual strain. Therefore, typically one to five strains were tested at the same time against one positive (n = 3) and one negative (n = 3) control, depending on the availability of larvae. Overall, 70 positive and 70 negative controls were conducted during mono- and multispecies biofilm settlement experiments. After Petri dishes had been prepared, they were placed outside in the shade to avoid high solar radiation. The temperature within the Petri dishes varied depending on the outside temperature (between 25 and 31 °C) as no incubator was available at the time. Preliminary experiments showed that the most larval settlement behavior happens within the first 48 h. Using a stereomicroscope, larval reactions were differentiated into four categories: (i) swimming larvae (no reaction), (ii) attachment, (iii) metamorphosis, and (iv) settlement (attachment and metamorphosis) (Fig. 1). The last three reactions (ii–iv) were considered as general settlement behavior. Coral larvae were designated “attached” if soft water movements by a pipette motion (ca. 0.5 ml s−1, 1 mm diameter and in 5 mm distance to the larvae) could not detach them. Furthermore, larvae were designated “metamorphosed” as soon as they developed an oral disk and clearly visible septae. It was observed that larvae can perform metamorphosis prior to attachment; however, this phenomenon did not lead to successful settlement. After metamorphosis, young polyps show typical “flower-like” forms. Finally, larvae were designated “settled” if both reactions (ii) and (iii) described above happened. Hereafter, settlement behavior data are given in mean + standard error (SE). Figure 1a–d displays the four stages: swimming (no reaction), attachment, metamorphosis, and settlement in larvae of L. purpurea. Coral larvae settlement behavior was assessed after 48 h. Since settlement behavior data were not normally distributed, all data points were tested against the negative controls for significance (except for those whose three replicates of one treatment were all 0) using a nonparametric Kruskal–Wallis test followed by a pairwise Dunn’s test with Bonferroni correction to perform multiple comparisons. Taking into account that one negative control was performed for multiple strains, settlement behavior data of each biofilm tested in triplicate, including multispecies biofilms, were compared to the sum of negative controls with n = 70.
Results
The isolation of bacteria from the CCA Hydrolithon reinboldii resulted in 56 strains. These strains belonged to six classes and 18 families (see Fig. 2 and Table 2). The culturable bacteria associated with H. reinboldii were dominated by Gammaproteobacteria (29 strains), followed by Actinobacteria (10 strains) and Alphaproteobacteria (9 strains). The bacterial classes Bacilli, Flavobacteria, and Cytophagia were only weakly represented with solely 5, 2, and 1 culturable strains, respectively. At family level (see Fig. 2), the CCA-associated bacterial community was dominated by Vibrionaceae and Rhodobacteraceae (both 9 strains), followed by Pseudoalteromonadaceae (7 strains), Alteromonadaceae and Micrococcaceae (5 strains, respectively), Pseudomonadaceae (4 strains), and Bacillales incertae sedis (3 strains). The remaining 14 bacterial strains were assigned to 11 families. Of all 56 isolates, 49 strains could be identified on species level (see Table 2). Although a broad range of different media with several supplements were used for the bacterial isolation experiments, we did not observe a clear benefit from certain media, treatments, or supplements such as CCA extract or antibiotics, except for MB media. Almost all strains used in the present study were exclusively isolated from MB plates.
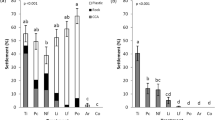
All 56 isolated microbial strains were used for the settlement behavior experiments. In most cases, there was large variation in induction by the strains. The six isolates #1792 (Pseudovibrio denitrificans; 73.3 ± 13.3%), #1678 (Acinetobacter pittii; 53.3 ± 24.0%), #1633 (Pseudoalteromonas phenolica; 46.7 ± 6.7%), #1772 (Marine bacterium LMG1; 46.7 ± 26.7%), #1721 (Microbulbifer variabilis; 40 ± 23.1%), and #1783 (Pseudoalteromonas rubra; 33.3 ± 6.7%) mostly induced settlement behavior at moderate (≥ 20%) to high (≥ 40%) and significant (except for #1721) levels in L. purpurea larvae (see Fig. 3 and Table 2). Another six strains induced moderate mean settlement behavior responses above 20% but mostly had large variations and were therefore not significantly different from the negative control. In another 16 strains, some settlement behavior was observed, although at low mean rates (below 20%) with large variations. The remaining 28 bacterial strains did not induce any settlement behavior at all. Attachment without metamorphosis was observed only in a few instances (0.8 ± 3.1% of all coral larvae), while metamorphosis in the water column was observed more frequently but again at very low levels (3.0 ± 5.8% of all tested larvae in response to biofilms). Interestingly, the latter phenomenon was only recorded in response to five bacterial strains (strains #1634, #1810, #1693, #1694, and #1701). Positive and negative controls resulted in 82.6 ± 2.2% and 0.6 ± 0.9% settlement behavior and provided a robust control setup throughout the larval experiments.
Settlement behavior of Leptastrea purpurea larvae in response to monospecific biofilms of bacterial strains isolated from Hydrolithon reinboldii. Legend: numbers behind class names represent the quantity of inductive strains. Numbers in brackets represent the quantity of tested strains within a class. Colored bars represent mean settlement behavior of coral larvae (n = 3) and standard errors ( ±). CCA served as positive control (n = 70) while FSW (filtrated seawater) served as negative control (n = 70). Percentages of swimming and dead larvae are displayed in light and dark gray, respectively. Bacterial strains inducing at significant levels were marked with *(p < 0.05)
Interestingly, coral larvae exposed to monospecific biofilms of the two Pseudoalteromonas strains #1784 (P. luteoviolacea, data not shown in Fig. 3) and #1783 (P. rubra) as well as strain #1772 (Marine bacterium LMG1) showed another reaction besides settlement behavior. Larvae exposed to strain #1784 became immobile and in some cases disintegrated after 24 h. After 48 h, all tested coral larvae had died and fully disintegrated. The bacterial strains #1772 and #1783 induced similar mortalities in L. purpurea larvae. Within the first 48 h, exposure to each of the two strains resulted in at least 20% mortality. After 72 h of exposure to strain #1772 or strain #1783 mortalities had increased to 40 and 100%, respectively. In comparison, the negative controls remained unaffected and alive after 72 h. Because of the latter observations, additional experiments were performed comparing larval mortalities after exposure to multispecies biofilms with or without strain #1772 (see Fig. 4). Strains that were observed to cause strong settlement behavior (above 40% on average) in coral larvae were used for the experiment. For this purpose, strains #1792, #1678, #1633, #1772, and #1721 were selected to create a multispecies biofilm. Coral larvae exposed to this multispecies biofilms all died within 24 h (see Fig. 4, mix + #1772). In contrast, multispecies biofilms composed of the same strains but without #1772 resulted in high levels of settlement behavior (93.2 ± 6.6% after 48 h) and no mortality, thereby surpassing the positive control.
Effect of multispecies biofilms on the settlement behavior of Leptastrea purpurea larvae. Multispecies biofilms were composed of strains #1792 (Pseudovibrio denitrificans), #1678 (Acinetobacter pittii), #1633 (Pseudoalteromonas phenolica), and #1721 (Microbulbifer variabilis) either with or without strain #1772 (Marine bacterium LMG1). Black bars represent mean settlement behavior of coral larvae (n = 3) and standard errors ( ±). CCA served as positive control (n = 70) while FSW (filtrated seawater) served as negative control (n = 70). Percentages of swimming and dead larvae are displayed in light and dark gray, respectively. Inducing activities at significant levels were marked with *(p < 0.05)
The bacterial communities from CCA fragments (n = 3), CCA cotton swabs (n = 3) and for comparison from sediment samples (n = 3) and a negative control (n = 1) were investigated by 16S rRNA gene amplicon sequencing. Processing of the 16S reads with Mothur yielded 134,467 high-quality reads, with an average sequencing depth of 13,447 ± 14,324 reads. One CCA cotton swab sample had relatively low sequencing depth (129 high-quality reads), and it was therefore removed from further analysis. Subsequently, contamination was reduced by subtraction of OTUs from the negative control from the biological samples. Finally, the remaining samples were rarefied to an even sequencing depth of 2640 reads. Figure 5 shows that the bacterial communities of all samples were dominated by Proteobacteria. In CCA swabs, Gammaproteobacteria accounted for 52.4 ± 18.5% on average, followed by Alphaproteobacteria with 8.6 ± 0.1%. Beta-, Delta-, and Epsilonproteobacteria averaged ≤ 5%. In contrast, the community of Proteobacteria in CCA fragments was dominated by lpha- (25.5 ± 3.4%) and Deltaproteobacteria (18.1 ± 6.5%), while Gammaproteobacteria accounted for only 11.6 ± 0.5% and Beta- as well as Epsilonproteobacteria stayed below 1%. The Proteobacteria community of the sediment samples was dominated by Gamma-, Delta-, and Alphaproteobacteria (25.5 ± 1.7%, 11.6 ± 2.2%, and 10.7 ± 0.9%, respectively) with Epsilon- and Betaproteobacteria being below 1% too. However, Fig. 5 shows that the phyla Planctomycetes and Bacteroidetes were present in all samples. With some variation, the phylum Cyanobacteria was found in all samples as well, although only in low abundance in swab 1. Other phyla that were found in all samples include Acidobacteria, Chloroflexi, Verrucomicrobia, Tenericutes, and Actinobacteria as well as unclassified bacteria. Further phyla that were found in different samples only accounted with minor amounts (< 1%) and were therefore included as “Other phyla.” Comparing the genus-level annotations of our CCA isolates from sanger sequencing with the genus-level annotation of the OTUs from Illumina MiSeq sequencing revealed that 11 of the 28 culturable genera were also detected in samples of either sediment, CCA fragments and/or CCA surface swabs (see Table 2). The two culturable genera Mycobacterium and Pseudomonas were exclusively detected in the sediment, the genera Aquimarina and Paracoccus were exclusively detected in CCA surface swabs, and no genus was exclusively detected in CCA fragments. Of the five isolates that significantly induced settlement behavior in coral larvae at high and moderate levels, only the two genera Pseudoalteromonas (#1633 and #1783) and Acinetobacter (#1678) were detected in the Illumina MiSeq samples, including CCA fragments, CCA swabs, and sediment.
Discussion
In this study, fragments of the CCA Hydrolithon reinboldii and swabs from the CCA surface led to the isolation of 56 microbial strains from 6 different bacterial classes and 18 families (see Table 2). Of the 56 isolates only in five strains settlement behavior of L. purpurea larvae was found to be statistically significant: strain #1792 (Pseudovibrio denitrificans), #1678 (Acinetobacter pittii), #1633 (Pseudoalteromonas phenolica), #1772 (Marine bacterium LMG1), and #1783 (Pseudoalteromonas rubra). That is, the inductive effects of bacterial isolates on coral larvae were found to be strain specific rather than exclusively linked to certain higher taxonomic levels, as, for example, the five isolates #1792, #1678, #1633, #1772, and #1783 that induced the highest and significant settlement behavior rates in this study belonged to five different species, three different genera, and two classes of bacteria. Similar results were reported by Tran and Hadfield (2011), who recorded settlement in larvae of the scleractinian coral Pocillopora damicornis in response to monospecific biofilms. They also concluded that there was no correlation between inductive capacities and phylogenetic relationships.
In this work, strain #1792 induced settlement behavior in L. purpurea larvae at the highest level compared to the other isolates. This genus has been isolated from a great variety of marine sources (Romano 2018 and referenced therein) and has shown to produce versatile secondary metabolites with several activities, including antibacterial (Penesyan et al. 2011) and cytotoxic (Rodrigues et al. 2017) effects. Interestingly, diindol-3-ylmethanes, isolated from Pseudovibrio denitrificans, showed moderate to strong inhibitory effects against larval settlement of the barnacle Balanus amphitrite (Wang et al. 2015). To the best of our knowledge, this is the first report of Pseudovibrio denitrificans inducing settlement behavior in larvae of a scleractinian coral. Hence, future studies on the isolate #1792 belonging the genus Pseudovibrio and its putative settlement cues for coral larvae are of interest. In contrast, strain #1678 belongs to a genus, namely Acinetobacter, which is known to induce moderate settlement in larvae of the scleractinian coral Pocillopora damicornis (Tran and Hadfield 2011). Unfortunately, the settlement inducing activity of this genus has not yet been linked to a certain chemical compound. Instead, a not further determined strain of Acinetobacter has been reported to produce the bromoindole derivative 6-bromoindole-3-carbaldehyde that inhibited the attachment of barnacle larvae (Olguin-Uribe et al. 1997). Strains #1633 (Pseudoalteromonas phenolica) and #1783 (P. rubra) belong to the genus Pseudoalteromonas and induced settlement behavior in L. purpurea larvae at high or moderate and statistically significant levels. A species of this genus, isolated from the CCA Hydrolithon onkodes, was reported to induce significant levels of metamorphosis in larvae of the corals Acropora willisae and A. millepora (Negri et al. 2001). Furthermore, both Tebben et al. (2011) and Sneed et al. (2014) isolated the morphogen tetrabromopyrrole from a CCA-associated Pseudoalteromonas strain. This cue induced metamorphosis and even settlement of several coral larvae, including the brooding coral Porites astreoides as well as the spawning corals Orbicella franksi, A. palmata and A. millepora. Noteworthy, strain #1772 had been identified as Marine bacterium LMG1 but shared a similarity of 99% with the species Pseudoalteromonas rubra using BLASTn and the genetic sequence database GenBank. Thus, further studies on the metabolomes of the two inductive Pseudoalteromonas strains #1633 and #1783 as well as on strain #1772 (Marine bacterium LMG1) will be conducted and may reveal the presence of tetrabromopyrrole and/or other possible inducers.
In general, rates of coral settlement behavior varied among all categories (strong, moderate, and weak) and isolates in the preset study, resulting in the majority of induction effects remaining insignificant compared to the negative controls. The problem of not showing many statistically significant results, in spite of high settlement behavior rates, were the high standard errors, ultimately leading to a high number of statistically insignificant results. A reason for the observed high variability among replicates in each treatment might have been the low numbers of replicates (n = 3) and larvae per replicate (5), due to limitation of available larvae specimen in general. Differences in the inductive activity among bacterial strains again might be a result of the non-standardized biofilms, as cell numbers for, e.g., inoculation were not counted. Although all liquid pre-cultures were incubated for the same time (48 h) and biofilms were clearly visible on the coverslips, there might have been large variations in the growth rates among the isolates. As a result, bacterial overgrowth and associated toxic by-products as well as the sheer density of some bacterial biofilms might have affected L. purpurea larvae and contributed to the overall variability of larval settlement behavior in response to the different strains. It is noteworthy that all larvae that did not settle in response to monospecific biofilms of strains #1772 and #1783 (P. rubra) died soon after. Separate from settlement compounds like tetrabromopyrrole, Pseudoalteromonas spp. are prolific producers of potent natural products such as antibacterials and cytotoxic compounds (Bowman 2007 and referenced therein; Offret et al. 2016 and referenced therein), some of which could have negative effects on coral larvae settlement if produced. Future experiments testing the effect of different growth phases of inductive (and to some extent toxic) isolates on the settlement behavior of coral larvae may help to identify possible settlement compounds and reveal their positive and/or negative role in this metamorphic process. In addition to toxic effects, #1634, #1693, #1694, #1701, and #1810 were the only strains where we observed metamorphosis without prior attachment in L. purpurea larvae. Studies on the settlement of marine invertebrate larvae using neurotransmitters showed that attachment and metamorphosis must not be linked and therefore might require different compounds (Iwao et al. 2002; Grasso et al. 2011; Moeller et al. 2019). That is, the aforementioned strains might have produced metabolites, likely water-soluble, which triggered solely metamorphosis but not attachment in coral larvae.
Another observation in this study was that every tested bacterial monospecies biofilm induced lower settlement behavior rates than the positive controls, being CCA (82.6% ± 18.8). Even though CCA (Morse et al. 1988; Price 2010; Ritson-Williams et al. 2014) and organic extracts of CCA (Heyward and Negri 1999; Harrington et al. 2004; Kitamura et al. 2009) are known to be potent settlement inducers, the present study joins a series of publications that display the dynamic ability of CCA-associated bacteria to induce settlement behavior in scleractinian corals (Negri et al. 2001; Tran and Hadfield 2011). We could further demonstrate that this dynamic ability can be increased by combining inductive strains to create multispecies biofilms. Larvae in response to multispecies biofilms consisting of strains #1792, #1678, #1633, and #1721 (Microbulbifer variabilis) had a higher mean settlement behavior response than any of the monospecies biofilms or the positive controls. The formation of biofilms, both mono- and particularly multispecies ones, can be advantageous for bacteria because it lays the foundation for cell-to-cell signaling and communication (Stoodley et al. 2002; Pasmore and Costerton 2003). Costerton and colleagues (1995) showed that multispecies biofilms have greater variability in their three-dimensional structure than monospecific biofilms, which may be an explanation for the here observed increased settlement behavior rates. Moreover, co-cultivation of bacteria increases the chemical diversity (Marmann et al. 2014 and referenced therein) and may have led to the production of more or other settlement behavior inducing or enhancing compounds. The addition of #1772 to multispecies biofilms resulted in a total loss of inducing activity as all L. purpurea larvae died and dissolved. This particular result can have different reasons: (i) strain #1772 overgrew the other strains; (ii) interactions between the five strains in which #1772 reduced the settlement behavior inducing activity of the four remaining strains (e.g., by competition and the production of toxic compounds); (iii) a combination of (i) and (ii). However, the interactions between bacteria within multispecies biofilms remain complex but might be a critical factor for the settlement success of coral larvae.
The CCA-associated bacterial community was analyzed in order to gain further insights into the role of the inductive and toxic bacteria in this niche. In general, CCA fragments and surface swabs hosted distinct bacteriomes that largely differed from the sediment samples (see PCoA plot in Fig. S1, online resource 1) but also from another (the two none-to-weak inducing genera Aquimarina and Paracoccus were only found in CCA surface swabs). The composition of the surface-associated culture-independent bacterial community of H. reinboldii (CCA swabs) was similar to previously reported results (Sneed et al. 2015). The short length of the Illumina MiSeq reads permitted confident taxonomic assignment at the genus level. Therefore, the comparisons between culture-dependent and -independent bacterial communities in this study were conducted at genus level. 17 of the 28 culturable genera could not be found in the Illumina MiSeq samples, but might have been below the detection threshold. This suggests that these genera may not be dominant members of the CCA-associated bacterial community. With regard to their ecological significance, one can only draw speculative conclusions based on the presence of certain genera, but more substantiated conclusions based on the absence or low abundance. For example, strain #1792 (Pseudovibrio), which was not detected in the Illumina MiSeq samples, induced settlement behavior in L. purpurea larvae at the highest level among all isolates. Hence, undetected genera might be relevant but their precise role in the settlement process of L. purpurea remains to be deciphered. In contrast, both settlement behavior inducing genera Pseudoalteromonas (#1633 and #1783), which has shown toxic effects upon larvae as well, and Acinetobacter (#1678) were found in Illumina MiSeq samples of CCA fragments and CCA surface swabs. The genus Pseudoalteromonas was also found in the sediment samples, which is of no surprise as it consists of obligate marine bacteria that are highly abundant in the marine environment and have been isolated from various marine sources, including (deep-sea) sediments, seawater, sea ice, but also macroorganisms such as algae, sponges, and mussels (Chan et al. 1978; Bowman 1998; 2007 and referenced therein; Park et al. 2005; Qin et al. 2011). However, although Illumina MiSeq sequence analyses have indicated the presence of settlement inducing genera on CCA (e.g., Pseudoalteromonas and Acinetobacter), the ecological significance of the here discussed inductive and toxic bacterial strains continues to be determined. For a higher taxonomic resolution, we suggest applying alternative sequencing technologies such as metagenomic shotgun sequencing or Pacific Biosciences (PacBio) single-molecule real-time sequencing (Earl et al. 2018). In addition, fluorescence-based imaging techniques such as FISH (fluorescence in situ hybridization) or imaging mass spectrometry such as DESI (desorption electrospray ionization) or MALDI (matrix-assisted laser desorption/ionization) may help to unequivocally prove the occurrence of settlement inducing strains and estimate their ecological relevance, i.e., by quantifying cell numbers, analyzing the role of their bioactive compounds, and screening for bacterial fingerprint compounds.
This study gave new insights into the microbial community associated with the CCA H. reinboldii and the potential of specific CCA-associated strains to induce settlement behavior in larvae of stony corals. Our findings may help to provide additional approaches for reef restoration projects of threatened coral reefs. Furthermore, the results of this study highlight the putatively large variety of settlement cues among bacteria and further emphasized the ambiguous connection between the genus Pseudoalteromonas and coral larvae settlement. Moreover, this study demonstrated that only a few other H. reinboldii-associated bacteria held (significant) inductive capacities for the settlement of L. purpurea larvae. Since many of the isolated species are known for being potent producers of versatile bioactive molecules, possible settlement cues may not be restricted to a single class of chemical compounds. Thus, the true nature of the chemical and/or physical interactions between biofilms and coral larvae remains yet to be deciphered. Further investigations on the induction mechanisms of a putative settlement cue will be intensified to find out if larvae respond to a rather insoluble surface-bound or to a soluble water-borne compound.
References
Ainsworth TD, Heron SF, Ortiz JC, Mumby PJ, Grech A, Ogawa D, Eakin CM, Leggat W (2016) Climate change disables coral bleaching protection on the great barrier reef. Sci 352:338–342
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Apprill A, McNally S, Parsons R, Weber L (2015) Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137
Barbier EB, Hacker SD, Kennedy C, Koch EW, Stier AC, Silliman BR (2011) The value of estuarine and coastal ecosystem services. Ecol Monogr 81:169–193
Bellwood DR, Hughes TP, Folke C, Nyström M (2004) Confronting the coral reef crisis. Nat 429:827–833
Bowman JP (1998) Pseudoalteromonas prydzensissp. nov., a psychro-trophic, halotolerant bacterium from Antarctic sea ice. Int J Syst Bact 48:1037–1041
Bowman JP (2007) Bioactive compound synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Mar Drugs 5:220–241
Chan KY, Baumann L, Garza MM, Baumann P (1978) Two new species of Alteromonas: Alteromonas espejiana and Alteromonas undina. Int J Syst Bact 28:217–222
Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM (1995) Microbial biofilms. Annu Rev Microbiol 49:711–745
Dobretsov S, Qian P-Y (2006) Facilitation and inhibition of larval attachment of the bryozoan Bugula neritina in association with mono-species and multi-species biofilms. J Exp Mar Bio Ecol 333:263–274
Earl JP, Adappa ND, Krol J, Bhat AS, Balashov S, Ehrlich RL, Palmer JN, Workman AD, Blasetti M, Sen B, Hammond J, Cohen NA, Ehrlich GD, Mell JC (2018) Species-level bacterial community profiling of the healthy sinonasal microbiome using Pacific Biosciences sequencing of full-length 16S rRNA genes. Microb 6:190. https://doi.org/10.1186/s40168-018-0569-2
Edmunds PJ (2004) Juvenile coral population dynamics track rising seawater temperature on a Caribbean reef. Mar Ecol Prog Ser 269:111–119
Erwin PM, Song B, Szmant AM (2008) Settlement behavior of Acropora palmata planulae: effects of biofilm age and crustose coralline algal cover. Proc 11th Int Coral Reef Symp 24:1219–1224
Grasso LC, Negri AP, Fôret S, Saint R, Hayward DC, Miller DJ, Ball EE (2011) The biology of coral metamorphosis: molecular responses of larvae to inducers of settlement and metamorphosis. Dev Biol 353:411–419
Hadfield MG (2011) Biofilms and marine invertebrate larvae: what bacteria produce that larvae use to choose settlement sites. Annu Rev Mar Sci 3:453–470
Harrington L, Fabricius K, De’Ath G, Negri A, (2004) Recognition and selection of settlement substrata determine post-settlement survival in corals. Ecol 85:3428–3437
Heron SF, Maynard JA, van Hooidonk R, Eakin CM (2016) Warming trends and bleaching stress of the world’s coral reefs 1985–2012. Sci Rep 6:38402. https://doi.org/10.1038/srep38402
Heyward AJ, Negri AP (1999) Natural inducers for coral larval metamorphosis. Coral Reefs 18:273–279
Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, Gomez E, Harvell CD, Sale PF, Edwards AJ, Caldeira K, Knowlton N, Eakin CM, Iglesias-Prieto R, Muthiga N, Bradbury RH, Dubi A, Hatziolos ME (2007) Coral reefs under rapid climate change and ocean acidification. Sci 318:1737–1742
Iwao K, Fujisawa T, Hatta M (2002) A cnidarian neuropeptide of the GLWamide family induces metamorphosis of reef-building corals in the genus Acropora. Coral Reefs 21:127–129
Jiang H, Dong H, Zhang G, Yu B, Chapman LR, Fields MW (2006) Microbial diversity in water and sediment of lake chaka, an athalassohaline lake in northwestern China. Appl Environ Microbiol 72:3832–3845
Johnson CR, Sutton DC, Olson RR, Giddins R (1991) Settlement of crown-of-thorns starfish: role of bacteria on surfaces of coralline algae and a hypothesis for deepwater recruitment. Mar Ecol Prog Ser 71:143–162
Kitamura M, Schupp PJ, Nakano Y, Uemura D (2009) Luminaolide, a novel metamorphosis-enhancing macrodiolide for scleractinian coral larvae from crustose coralline algae. Tetr Lett 50:6606–6609
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl Environ Microbiol 79:5112–5120
Marmann A, Aly AH, Lin W, Wang B, Proksch P (2014) Co-cultivation-a powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar Drugs 12:1043–1065
Moeller M, Nietzer S, Schils T, Schupp PJ (2017) Low sediment loads affect survival of coral recruits: the first weeks are crucial. Coral Reefs 36:39–49
Moeller M, Nietzer S, Schupp PJ (2019) Neuroactive compounds induce larval settlement in the scleractinian coral Leptastrea purpurea. Sci Rep 9:2291. https://doi.org/10.1038/s41598-019-38794-2
Morse DE, Morse ANC (1991) Enzymatic characterization of the morphogen recognized by Agaricia humilis (scleractinian coral) larvae. Biol Bull 181:104–122
Morse DE, Hooker N, Morse ANC, Jensen RA (1988) Control of larval metamorphosis and recruitment in sympatric agariciid corals. J Exp Mar Biol Ecol 116:193–217
Negri AP, Webster NS, Hill RT, Heyward AJ (2001) Metamorphosis of broadcast spawning corals in response to bacteria isolated from crustose algae. Mar Ecol Prog Ser 223:121–131
Negri AP, Marshall PA, Heyward AJ (2007) Differing effects of thermal stress on coral fertilization and early embryogenesis in four Indo Pacific species. Coral Reefs 26:759–763
Nietzer S, Moeller M, Kitamura M, Schupp PJ (2018) Coral larvae every day: Leptastrea purpurea, a brooding species that could accelerate coral research. Front Mar Sci 5:466. https://doi.org/10.3389/fmars.2018.00466
Offret C, Desriac F, Le Chevalier P, Mounier J, Jégou FY (2016) Spotlight on antimicrobial metabolites from the marine bacteria Pseudoalteromonas: chemodiversity and ecological significance. Mar Drugs 14:129. https://doi.org/10.3390/md14070129
Olguin-Uribe G, Abou-Mansour E, Boulander A, Débard H, Francisco C, Combaut G (1997) 6-bromoindole-3-carbaldehyde, from an Acinetobacter sp. bacterium associated with the ascidian Stomozoa murrayi. J Chem Ecol 30:2507–2521
Parada AE, Needham DM, Fuhrman JA (2016) Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414
Park Y-D, Baik KS, Yi H, Bae KS, Chun J (2005) Pseudoalteromonas byunsanensis sp. nov., isolated from tidal flat sediment in Korea. Int J Syst Evol Microbiol 55:2519–2523
Pasmore M, Costerton JW (2003) Biofilms, bacterial signaling, and their ties to marine biology. J Ind Microbiol Biotechnol 30:407–413
Pauly D, Christensen V, Guénette S, Pitcher TJ, Sumaila R, Walters CJ, Watson R, Zeller D (2002) Towards sustainability in world fisheries. Nat 418:689–695
Pawlik JR (1992) Chemical ecology of the settlement of benthic marine invertebrates. Oceanogr Mar Biol Annu Rev 30:273–335
Penesyan A, Tebben J, Lee M, Thomas T, Kjelleberg S, Harder T, Egan S (2011) Identification of the antibacterial compound produced by the marine epiphytic bacterium Pseudovibrio sp. D323 and related sponge-associated bacteria. Mar Drugs 9:1391–1402
Price N (2010) Habitat selection, facilitation, and biotic settlement cues affect distribution and performance of coral recruits in French Polynesia. Oecol 163:747–758
Qin Q-L, Li Y, Zhang Y-J, Zhou Z-M, Zhang W-X, Chen X-L, Zhang X-Y, Zhou B-C, Wang L, Zhang Y-Z (2011) Comparative genomics reveals a deep-sea sediment-adapted life style of Pseudoalteromonas sp. SM9913. ISME J 5:274–284
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The silva ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596
Ritson-Williams R, Paul VJ, Arnold SN, Steneck RS (2010) Larval settlement preferences and post-settlement survival of the threatened caribbean corals Acropora palmata and A. cervicornis. Coral Reefs 29:71–81
Ritson-Williams R, Arnold SN, Paul VJ, Steneck RS (2014) Larval settlement preferences of Acropora palmata and Montastraea faveolata in response to diverse red algae. Coral Reefs 33:59–66
Ritson-Williams R, Arnold SN, Fogarty ND, Steneck RS, Vermeij MJA, Paul VJ (2009) New perspectives on ecological mechanisms affecting coral recruitment on reefs. Smithson Contrib Mar Sci 38:437–457
Rodrigues AMS, Rohée C, Fabre T, Batailler N, Sautel F, Carletti I, Nogues S, Suzuki MT, Stien D (2017) Cytotoxic indole alkaloids from Pseudovibrio denitrificans BBCC725. Tetr Lett 58:3172–3173
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: a versatile open source tool for metagenomics. Peer J 4:e2584. https://doi.org/10.7717/peerj.2584
Romano S (2018) Ecology and biotechnological potential of bacteria belonging to the genus Pseudovibrio. Appl Environ Microbiol 84:e02516-e2517. https://doi.org/10.1128/AEM.02516-17
Sneed JM, Ritson-Williams R, Paul VJ (2015) Crustose coralline algal species host distinct bacterial assemblages on their surfaces. ISME J 9:2527–2536
Sneed JM, Sharp KH, Ritchie KB, Paul VJ (2014) The chemical cue tetrabromopyrrole from a biofilm bacterium induces settlement of multiple caribbean corals. Proc R Soc B 281:20133086. https://doi.org/10.1098/rspb.2013.3086
Stoodley P, Sauer K, Davies DG, Costerton JW (2002) Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209
Tebben J, Tapiolas DM, Motti CA, Abrego D, Negri AP, Blackall LL, Steinberg PD, Harder T (2011) Induction of larval metamorphosis of the coral Acropora millepora by tetrabromopyrrole isolated from a Pseudoalteromonas bacterium. PLoS ONE 6:e19082. https://doi.org/10.1371/journal.pone.0019082
Tebben J, Motti CA, Siboni N, Tapiolas DM, Negri AP, Schupp PJ, Kitamura M, Hatta M, Steinberg PD, Harder T (2015) Chemical mediation of coral larval settlement by crustose coralline algae. Sci Rep 5:10803. https://doi.org/10.1038/srep10803
Thiyagarajan V, Lau SCK, Cheung SCK, Qian P-Y (2006) Cypris habitat selection facilitated by microbial films influences the vertical distribution of subtidal barnacle Balanus trigonus. Microb Ecol 51:431–440
Tran C, Hadfield MG (2011) Larvae of Pocillopora damicornis (anthozoa) settle and metamorphose in response to surface-biofilm bacteria. Mar Ecol Prog Ser 433:85–96
Wang K-L, Xu Y, Lu L, Li Y, Han Z, Zhang J, Shao C-L, Wang C-Y, Qian P-Y (2015) Low-toxicity diindol-3-ylmethanes as potent antifouling compounds. Mar Biotechnol 17:624–632
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Webster NS, Smith LD, Heyward AJ, Watts JEM, Webb RI, Blackall LL, Negri AP (2004) Metamorphosis of a scleractinian coral in response to microbial biofilms. Appl Environ Microbiol 70:1213–1221
Acknowledgements
This study was carried out in the framework of the PhD research training group “The Ecology of Molecules” (EcoMol) supported by the Lower Saxony Ministry for Science and Culture. We also thank the UOG Marine Lab and staff for support during the bacterial isolation and settlement behavior assays.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
PJ Schupp and M Moeller designed the project. M Moeller and S Nietzer performed the initial isolation of bacteria. M Moeller performed growth experiments and settlement behavior assays. D Versluis, PJ Schupp and L-E Petersen generated and processed amplicon data. L-E Petersen wrote the manuscript. All authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Topic Editor Dr. Anastazia Teresa Banaszak
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Petersen, LE., Moeller, M., Versluis, D. et al. Mono- and multispecies biofilms from a crustose coralline alga induce settlement in the scleractinian coral Leptastrea purpurea. Coral Reefs 40, 381–394 (2021). https://doi.org/10.1007/s00338-021-02062-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00338-021-02062-5