Abstract
Hordeum vulgare var. nudum (naked barley) is one of the oldest and most common cereals found from Neolithic Fennoscandia. After the Bronze Age, naked barley largely disappeared and was replaced by Hordeum vulgare var. vulgare (hulled barley) and other cereals. During the early 19th century, naked barley of Asian origins was reintroduced to Fennoscandia. In this study, we have genetically characterized samples of Fennoscandian landraces of naked barley which were preserved in gene banks and museum collections. The analyses show that the Fennoscandian naked barley can be split into three groups: First, naked two-row barley, with a likely origin in Asia; second, naked six-row barley, with a likely origin in the eastern Himalayas and introduced during the 19th century; third, naked six-row barley genetically related to the original Fennoscandian hulled barley. The results suggest that this last group represents the ancient form of naked barley, which was possibly introduced in the Neolithic. At that time both naked and hulled barleys were grown and enough gene flow probably occurred between these two subspecies to create a Fennoscandian barley that is genetically distinct, irrespective of whether it is hulled or naked. This hypothesis was further supported by genotyping of the Nud gene, which is responsible for the naked phenotype. All naked barleys which we studied contained the same mutation allele, nud1.a, thus showing that naked Fennoscandian barley arose by crossings between naked and hulled barley and not by new mutations of hulled barley.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The domestication of Hordeum vulgare L. (barley) began around 10,500 years bp (Zohary et al. 2012). Genetic analyses of wild barley and barley landraces (locally cultivated material maintained by farmers) suggest at least two domestication events, one in the fertile crescent of western Asia and one more eastern, possibly in the area of the Zagros mountains, east of the Iranian Plateau (Morrell and Clegg 2007; Saisho and Purugganan 2007). An additional centre of domestication has been suggested in Tibet (for example Dai et al. 2012), but was questioned by Zeng et al. (2018). A history of multiple domestication events is also supported by the genes btr1 and btr2 with independent causative mutations for a non-shattering rachis phenotype, and with selection events separated in time and space (Pourkheirandish et al. 2015). Analyses by Poets et al. (2015) further demonstrate that several populations of wild barley contributed to the gene pool of cultivated barley during the domestication process, resulting in environmentally adapted subpopulations.
Whereas wild barley is two-rowed and hulled, domesticated forms of barley also include six-row and naked (hull-less) types. The mutations causing these traits appear to be very old. Barley of the six-row type, with spikes with three fertile spikelets on each rachis node instead of one, appear in the archaeological record from 8,800 to 8,000 years bp (Helbæk 1959). At least four alleles (different forms) are known of the causative gene vrs1 which result in six-rowed plants (Komatsuda et al. 2007; Saisho et al. 2009), suggesting that the mutation has occurred repeatedly and then plants with this trait were selected. Naked barley, that is barley grains without adhering hulls (outer lemma and inner palea), have been discovered from around 8,000 years bp. Two naturally occurring alleles of the causative gene for nakedness nud have been identified. The first, and by far most common one, nud1.a, consists of a 17 kb deletion including the complete Nud gene (Taketa et al. 2004, 2008). The second one, nud1.g, has only been found in a few naked barleys from Tibet and is caused by a non-synonymous SNP (single nucleotide polymorphism) mutation (Yu et al. 2016).
Although naked barley occurs worldwide, the distribution of the hulled and naked forms varies greatly in time and space. At present, very little naked barley is grown in Europe and the Near East, whereas in eastern Asia and especially the Himalayan highlands, it can be very frequent (Takahashi 1955; Lister and Jones 2013). However, archaeological evidence clearly shows that naked barley used to be much more common in prehistoric Europe and the Near East. In a compilation of archaeological barley finds, Lister and Jones (2013) showed that for the Neolithic and Bronze Age around 40% of sites with barley finds had either naked barley or a mix of naked and hulled forms. For the Iron Age, the frequency of sites with naked barley drops markedly. This transition from naked to hulled barley is most pronounced in the Nordic countries. In southern Scandinavia naked barley was the main cereal grown during the Neolithic and Bronze Ages, but it was replaced by hulled barley during the transition between the late Bronze Age and pre-Roman Iron Age (Grabowski 2011). Regionally, as in parts of Jutland, Denmark, the cultivation of naked barley lasted well into the Roman Iron Age (Grabowski 2013). In Finland, finds are fewer, but they indicate that naked barley started to decrease during the late Iron Age (Vanhanen 2019).
The reasons for the transition from naked to hulled barley are not clear. The barley hull (husk) is inedible and must be removed if the grain is intended as food for humans. De-husking of hulled barley requires much work and as yields of naked and hulled barley are in the same range (Barabaschi et al. 2012), it seems peculiar that the Nordic prehistoric societies so clearly changed from naked to hulled barley. Grabowski (2011) suggests various reasons: Hulled barley can resist abiotic and biotic stress better, it responds better to manuring, it is good for animal feed, it is suitable for brewing beer, is more resistant to shattering when harvested with tools such as metal sickles and it stores better, as the husk provides protection against fungal or insect attacks. Whatever the cause, only occasional remains of naked barley have been found in the Nordic countries from after the Iron Age.
Whether the ancient Nordic naked barley completely disappeared is, however, unclear. Historical records from the early modern period mention naked barley sporadically and as a curiosity (Ahokas 2006; Leino 2017). In the 19th century a general belief was that seed taken from areas with a harsh climate would perform particularly well if grown in an area with better conditions (for example Lundequist 1855). Therefore, naked barley seed corn was imported from the highlands of Asia. The material was multiplied and distributed to farmers by the Royal Swedish Academy of Agriculture (Carling 1832; Leino 2017). Seed samples from the academy’s experimental fields in the 19th century have been preserved to this day (Fig. 1A). During the 20th century, Fennoscandian naked barley was collected by agronomists and gene banks (Fig. 1B-D; Åberg 1940; Ahokas 2006; Leino 2017). These collections are now curated as non-viable grain samples or as viable germplasm by the Nordic Genetic Resource Center (NordGen).
Examples of materials used in the analyses. A, sample of “Himmels-korn” (celestial barley) grown in 1894 on the experimental field in Stockhom (accession NM.0405688); B, Extant six-row naked barley (NGB 15229); C, sample of “Nakenkorn från Sörmark” (naked barley from Sörmark) from 1947 (Å128); D, Extant two-row naked barley (CIho 2512). Photos A, Peter Segemark; B, D, Matti W Leino; C, Peter Henriksson
The identity of the Fennoscandian germplasm of naked barley is, however, unknown. By genetically comparing extant material of naked and hulled barleys from Fennoscandia kept alive in gene banks and from museums and herbaria with that from the mountain regions of Central Asia, we have investigated whether the Fennoscandian germplasm is descended from the 19th century seed imports from Central Asia or is the remnant of an ancient population of Fennoscandian naked barley. In addition, we have screened the material for presence or absence of the 17 kb deletion of Nud to determine whether the nakedness in Fennoscandian barleys is caused by the same deletion allele as in most other naked barleys worldwide. Based on these analyses we propose a history for Fennoscandian naked barley.
Materials and methods
Plant materials
A total of 34 accessions were studied (Table 1). These were chosen to represent both naked and hulled barleys from both the Central Asian highlands of Afghanistan, Nepal and China, and from Fennoscandia (Denmark, Norway, Sweden and Finland). Both two- and six-row accessions were included. Accessions were obtained either from gene banks (the Nordic Genetic Resource Center, NordGen, prefix NGB and the National Small Grains Collection in the United States Department of Agriculture, USDA, prefix Clho) or from historical collections consisting of 19th century seed at the Nordic Museum (prefix NM) (Leino et al. 2009) and the Åberg collection from the 1950s at NordGen (prefix Å). The historical specimens consisted of non-viable harvest grain samples and some specimens also contained ear samples from which the row type (two or six) was determined. When ears were not present, the row type was determined from the ratio of straight to twisted grains according to Jacomet (2006). Extant viable accessions were grown and phenotyped for row type and hull characteristics.
DNA extraction
DNA was extracted from four to six individuals of each accession. Extant accessions from gene banks were germinated and DNA was extracted from the resulting leaves using the Qiagen DNeasy Plant Mini Prep Kit according to the manufacturer’s instructions. For the historical samples, DNA was extracted from the grains in a sterile hood that had never been used for work with modern material. Individual grains were crushed with sterile pliers and then the DNA was extracted using the MP Biomedicals FastPrep kit. Three rounds of homogenization were used and an extra hour of incubation was included after homogenization to increase the yield. Apart from these modifications, extractions were carried out according to the manufacturer’s instructions.
Nud genotyping
All individuals were genotyped for the 17 kb deletion in the Nud locus (nud1.a) described by Taketa et al. (2008). For extant specimens the Nud locus was genotyped with a PCR (polymerase chain reaction) assay according to Taketa et al. (2008), using two sets of primer combinations positioned either outside or inside the deletion. For the historical specimens where the degraded state of the DNA prevented amplification of the original PCR product, a novel forward genotyping primer, nud_short_F3, was used: 5’-ctacaaagccgtgggaatcg-3’. When used with the Taketa et al. (2008) tR2 (5’-gcggtcctttctttccagt-3’) primer on DNA not carrying the deletion (hulled phenotype) the resulting DNA fragment was 246 bp and when used with kR1 (5’-cctcaccacttaaccatgtctg-3’) on DNA carrying the 17 kb Nud deletion (naked phenotype) the resulting fragment was 168 bp. For each individual sample two separate PCRs were run with the two reverse primers tR2 and kR1 respectively, ensuring that each genotype was confirmed by a positive and a negative PCR result.
PCRs were run in a solution made up of 11.4 µl water, 2 µl Dreamtaq buffer, 0.4 µl 10 µM dNTPs (Thermo Scientific), 2 µl 1 µM of each primer and 0.2 µl DreamTaq polymerase (Thermo Scientific) with 2 µl template DNA added to each reaction. The PCR conditions were an initial denaturation at 95 °C for 2 min 30 s, 24 cycles of denaturation at 94 °C for 15 s, annealing at 56 °C for 40 s and extension at 72 °C for 40 s, followed by a final extension at 72 °C for 10 min.
SNP genotyping
The set of individuals was also genotyped for 85 SNP (single nucleotide polymorphism) markers (ESM Table S1). These were chosen from the genome-wide set of barley markers in the BOPA 1 array (Kota et al. 2008). For a subset of accessions, the causative SNP of the photoperiod response gene Ppd-H1 (SNP48, Jones et al. 2008) was included. Genotyping was carried out by LGC Genomics (Hoddeston, UK) using the KASP method (He et al. 2014).
Data analysis
The software Structure v. 2.2 (Pritchard et al. 2000) was used to investigate the dataset for population structure. As barley is self-fertilizing and largely homozygous, Structure was run in a haploid setting as suggested by Nordborg et al. (2005). It was run using the admixture model, with the length of the burn-in set to 20,000 and followed by 50,000 iterations to estimate parameters. Ten runs were done for each level of K ranging from 2 to 10. To assess the most appropriate number of clusters and to visualize the results, CLUMPP v. 1.1.2 (Jakobsson and Rosenberg 2007) was run with the Greedy algorithm for 4 < K < 6 and with the LargeKGreedy algorithm for K ≤ 6. The number of clusters best describing the data was evaluated from the CLUMPP H’ values and ΔK calculated according to Evanno et al. (2005). Results were visualized using Distruct v 1.1 (Rosenberg 2004).
R v. 4.1.2 was used to estimate within-accession diversity, to carry out principal component analysis (PCA) and to test for differences in the amount of within-accession genetic diversity between groups of accessions (R Core Team 2020). The hs function of the adegenet package was used to calculate HS as a measure of genetic diversity and the prcomp function was used for PCA. In PCA the frequency of each allele at each locus and accession was used. Diversity data was tested for normality with the Shapiro-Wilk test using the shapiro.test function.
Results
Genetic diversity
In total, genotyping data were analysed from 170 individuals which had been genotyped for 85 genetic markers (ESM Table S1). The success rate for all markers and also for all individuals was above 70% with an average of 96%. The average success rate in the historical material was 95%. Genetic diversity within each accession, calculated as the expected level of heterozygosity, ranged from 0 in four invariant (genetically identical) accessions to 0.220 in two hulled accessions from Afghanistan (ESM Table S1). No significant differences in the levels of genetic diversity within accessions could be detected between hulled and naked barley, nor between gene bank and museum material (Mann-Whitney U Test, p > 0.05).
Structure analysis
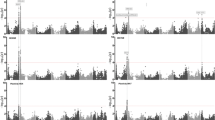
Both ΔK and H’ values for the Structure analysis suggested that the distribution of genetic diversity was best explained by a model with two clusters (ESM Table S2). At K = 2 both clusters contained both naked and hulled accessions (Fig. 2A). One cluster (dark blue) contained primarily Fennoscandian six-row barley, both hulled and naked. The other cluster (light blue) contained all forms of studied barley, both Asian and Fennoscandian, six- and two-row, and both hulled and naked forms. Some accessions belonged to both clusters, mainly Asian hulled accessions and two-rowed Fennoscandian hulled barley.
Results of Structure analyses of Asian and Fennoscandian barleys, A, with K = 2; B, with K = 9. Each vertical bar represents data from a single individual clustered in accessions separated by thin black lines. Different colours show the proportion of identity of that individual to each cluster (K) explained by the investigated model. The accessions are sorted by origin, hull type and row type. Historical (museum) accessions are marked with *
The second highest H’ value was obtained for K = 9 clusters (ESM Table S2). At this level of clustering, six- and two-row Fennoscandian hulled accessions formed individual clusters (light blue and red in Fig. 2B, respectively). Six-row Fennoscandian naked accessions fell into either of two clusters (light orange or dark green in Fig. 2B). The dark green cluster was also shared by naked six-row Asian accessions NGB 7320 and NM 214, and to a lesser extent by NGB 5094. Additionally, a few hulled barleys of both six-row and two-row Asian accessions were in the dark green cluster. The two-row Fennoscandian naked accessions formed a separate cluster (pink in Fig. 2B). Besides the dark green clustered naked six-row accessions, the Asian accessions showed a higher level of mixed clustering and mostly formed clusters separate from the Fennoscandian ones. The Asian naked six-row accession NGB 9305, however, mostly belonged to one of the Fennoscandian six-row naked clusters (light orange in Fig. 2B).
PCA
In a PCA of the genotype data, principal components 1, 2 and 3 explained 20.59, 15.48 and 14.60% of the variation respectively. The distribution of accessions along the three first principal components to a large extent reflected the results of the Structure analysis, both at K = 2 and K = 9. The groups observed in the PCA could not be explained by origin (Fig. 3A), hulledness (hulled or naked) (Fig. 3B) or row type (Fig. 3C) alone. Instead, all three aspects need to be considered together. The first cluster (positive end of PC1 in Fig. 3), separated from the other accessions along PC1, PC2 and PC3 was primarily made up of Fennoscandian six-row hulled and naked accessions, corresponding with the accessions forming the dark blue group at K = 2 of the Structure analysis. Within this cluster, hulled and naked barleys tended to form separate groups (Fig. 3B). The Asian accession NGB 9305 (naked 6-row) also belonged to this cluster but was located away from the hulled and naked subgroups (Fig. 3A). The second cluster (negative end of PC1 in Fig. 3) contained the remaining Asian and several Fennoscandian accessions. Among these, the naked Fennoscandian accessions were both two and six-rowed, while the Fennoscandian hulled accessions were two-row. Within the second cluster, hulled and naked accessions tended to cluster apart along PC3 (Fig. 3B), but no clear grouping could be observed for geographical origin or row type (Fig. 3A, C).
Nud genotyping
Screening for the nud1.a allele at the Nud locus was done by PCR genotyping according to the Taketa et al. (2008) assay. For the historical samples a new primer was developed, resulting in shorter PCR products which permit amplification of degraded DNA (Fig. 4). All individuals with a naked phenotype were confirmed to carry the 17 kb deletion, the nud1.a allele. This deletion was not found in any of the hulled individuals. The accession NGB 8872 phenotyped as a mix of hulled and naked individuals with specimens carrying the corresponding Nud and nud1.a alleles, respectively.
PCR genotyping results for the Nud locus in four individuals each of NGB 6292 (genebank, hulled), NGB 7320 (genebank, naked), Åberg353 (museum, naked) and NM.0405759 (museum, hulled) respectively. A, genotyping for nud.a1 (deletion = naked) allele using the kR1 primer with either wF2 (genebank individuals) or nud_short_F3 (museum individuals); B, genotyping for Nud (non-deletion = hulled) allele using the tR2 primer with either wF2 (genebank individuals) or nud_short_F3 (museum individuals). The accession NM.0405759 was not used for SNP genotyping and is only included to verify the functionality of the Nud genotyping of museum specimens
Discussion
The history of naked barley in Fennoscandia is shrouded in obscurity. Not only are the reasons for the decline of naked barley cultivation after the Bronze Age uncertain, but also the subsequent fate of the crop until the present day is not well known. During the 20th century several accessions of naked barley were collected in Fennoscandia and our genetic analyses of these samples give clues to the history of naked barley in the region.
Two introductions of naked barley to Fennoscandia
Structure and principal component analyses showed that naked Fennoscandian six-row barley belongs to two major groups (Figs. 2A and 3). One of the groups (dark blue in Fig. 2A) showed close similarities to hulled Fennoscandian six-row barley, while the other was more similar to both hulled and naked Asian barleys, both two and six-row. We suggest that these two genetically different groups represent two separate introduction events of naked barley into Fennoscandia.
The original introduction of six-row barley, possibly occurring during the Neolithic, was of both hulled and naked types. At this time both of them were grown extensively in parallel, possibly also mixed, although naked barley was more common (Grabowski 2011; Vanhanen et al. 2019). The fact that the dark blue cluster (Fig. 2A) includes both hulled and naked barley suggests that gene flow must have occurred between the two forms. Over time, a Fennoscandian barley developed that is genetically distinguishable from landraces from other regions, irrespective of whether it is hulled or naked. Further support for the ancient history of this group of naked barleys is given by the presence of the day-length non-responsive allele (ppd-H1) of the gene Ppd-H1 that controls flowering under long day conditions (Table 1). This allele is typical in landraces of barley from northern Europe and has been suggested to originate from the time of the first spread of agriculture in Europe (Lister et al. 2009; Jones et al. 2012). In contrast, all hulled and naked Asian barleys carry the responsive allele of Ppd-H1.
Previous investigations of hulled six-row Fennoscandian barley in this region have shown that it is highly distinguishable from all other European barleys, suggesting that it had a long period of adaptation and isolation (Aslan et al. 2015). The current study adds several naked Fennoscandian six-row barleys to this group. These (light orange in Fig. 2B) include both historical and extant accessions from Denmark, Sweden and Finland. Furthermore, principle components analysis shows that they are nearly homogeneous for the studied markers, despite their diverse origins in time and space (Fig. 3). These barleys also have low levels of genetic diversity within each accession (Table 1). A possible explanation for this is that the very limited cultivation of naked barley in Fennoscandia, at least since the 17th century (Ahokas 2006; Leino 2017), led to a reduction in genetic diversity. In addition, conservation of small samples in university collections and in gene banks might have reduced diversity within accessions even further (Hagenblad et al. 2012). Naked barley is very rare, although not completely absent, in archaeological and historical records from the late Iron Age to the 20th century (Viklund 1998; Grabowski 2011; Leino 2017). However, it seems to have been continuously propagated until the time when it was harvested for the historical seed collections and or deposited in the Nordic Genebank.
The second introduction of naked barley to Fennoscandia is reflected in the second group of Scandinavian naked barleys (light blue in Fig. 2A) and likely to be much more recent. This group carries the responsive Ppd-H1 allele (ESM Table S1) and shows more similarity to Asian barleys than to other Scandinavian landraces as would be expected from a more recent shared ancestry. In general, Asian barleys belong to a different genetic group than northern European (including Fennoscandian) ones (Muñoz-Amatriaín et al. 2014; Pasam et al. 2014). It is thus likely that this second group of naked Fennoscandian accessions originated from the 19th century importing of naked barley from Asia for the experimental field in Stockholm and distribution from there to the rest of Sweden (Carling 1832; Ahokas 2006; Leino 2017).
At higher levels of structuring, this group is split into two sub-groups corresponding to row type (dark green and pink in Fig. 2B). The group of six-row naked barleys (dark green in Fig. 2B) consists mostly of historical material and is very homogeneous. These barleys may possibly all originate from the same batch of seed corn which was imported from Vienna in 1824 as described by Carling (1832). The two-row naked barleys (NGB 8229 and CIho2512), and to some extent also the two-row hulled barleys, are genetically more similar to the Asian barleys than the six-row Fennoscandian barleys (Fig. 3). Comparisons with a more extensive range of naked Asian barleys could shed more light on the origin of the 19th century imports. The results also raise questions about the origin of Fennoscandian two-row hulled barley, a crop type considered to be only a few centuries old in Fennoscandia (Leino 2017).
The different genetic origins of the various samples are also reflected in the confusing nomenclature for naked barley. In Nordic languages, naked barley is frequently named with the prefixes himalais- (meaning dull sound), himmels- (meaning celestial, compare with the former Latin name ‘var. coeleste’) or Himalayan- (meaning originating in the Himalayas, compare with the former Latin name var. himalayense) (Ahokas 2006). Although the meanings of the three names are completely different, their spelling and pronunciation are similar, leading to misunderstandings. For example, among our accessions NM217 is named “himmelskorn” and NM1153 is named “Himalaya”, although both accessions are genetically identical and of Asian origin. In contrast, NM748 is named “Himalaya” and NGB 9305 is named “Näckte von Nepal” (Naked from Nepal) in spite of being genetically similar to the ancient Fennoscandian type. We thus caution against placing too much trust in accession names for geographical origin, at least in the case of Fennoscandian naked barley.
Fennoscandian naked barleys with the nud1.a allele
PCR genotyping confirmed the presence of the nud1.a allele in all individuals with the naked phenotype. Previous screenings of barley of world-wide origins have found that the nud1.a allele always occurs together with the naked grain phenotype (Taketa et al. 2004, 2008; Lei et al. 2020). Fennoscandian barleys have so far been poorly investigated, but this study confirms the presence of nud1.a in barley also from this part of the world. Thus, our results provide further support to Taketa et al. (2008) in suggesting an ancient origin of naked barley from a common ancestor. The only known exception so far is the nud1.g allele, which is found at low frequency among naked barleys from Tibet. Our study included several accessions from the central Asian highlands, but mainly from the more western parts. Together, it thus seems that the presence of the nud1.g allele is limited to a very restricted area in Tibet.
Conclusions
Our data support an ancient deletion mutation allele, nud1.a, that was present in naked barley when it, together with hulled barley, was introduced to Fennoscandia in the Neolithic period. Over time, gene flow between naked and hulled barleys in Fennoscandia has led to homogeneity between the two forms. During the 19th century both six- and two-row naked barleys were introduced from Asia to Fennoscandia. Both the ancient Fennoscandian six-row naked barley and the 19th century introductions have survived to the present day.
References
Åberg E (1940) The taxonomy and phylogeny of Hordeum L. sect. Cerealia ands. with special reference to Thibetan barleys. Dissertation, Uppsala University, Uppsala
Ahokas H (2006) Naked (nud) barley, Hordeum vulgare: a series of misunderstandings of its onomatopoetic name himalais- as himmels- (‘celestial’), himalayense and guimalaye (‘himalayan’) along with its migration. Kave, Helsinki
Aslan S, Forsberg NEG, Hagenblad J, Leino MW (2015) Molecular genotyping of historical barley landraces reveals novel candidate regions for local adaption. Crop Sci 55:2766–2776. https://doi.org/10.2135/cropsci2015.02.0119
Barabaschi D, Francia E, Tondelli A, Gianinetti A, Stanca AM, Pecchioni N (2012) Effect of the nud gene on grain yield in barley. Czech J Genet Plant Breed 48:10–22
Carling O (1832) Om himale-kornet och kornodling i allmänhet. Bredberg, Stockholm
Dai F, Nevo E, Wu D, Comadran J et al (2012) Tibet is one of the centers of domestication of cultivated barley. Proc Natl Acad Sci USA 109:16969–16973. https://doi.org/10.1073/pnas.1215265109
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Grabowski R (2011) Changes in cereal cultivation during the Iron Age in southern Sweden: a compilation and interpretation of the archaeobotanical material. Veget Hist Archaeobot 20:479–494. https://doi.org/10.1007/s00334-011-0283-5
Grabowski R (2013) Cereal cultivation in east-central Jutland during the Iron Age, 500 BC–AD 1100. Dan J Archaeol 2:164–196. https://doi.org/10.1080/21662282.2014.920127
Hagenblad J, Zie J, Leino MW (2012) Exploring the population genetics of genebank and historical landrace varieties. Genet Resour Crop Evol 59:1185–1199. https://doi.org/10.1007/s10722-011-9754-x
He C, Holme J, Anthony J (2014) SNP genotyping: the KASP assay. In: Fleury D, Whitford R (eds) Crop breeding: methods and protocols. Methods in Molecular Biology, vol 1145. Humana Press, New York, NY, pp 75–86. https://doi.org/10.1007/978-1-4939-0446-4
Helbæk H (1959) Domestication of food plants in the old world: joint efforts by botanists and archeologists illuminate the obscure history of plant domestication. Science 130:365–372. https://doi.org/10.1126/science.130.3372.365
Jacomet S (2006) Identification of cereal remains from archaeological sites, 2nd edn. IPAS, Basel University, Basel
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806https://doi.org/10.1093/bioinformatics/btm233
Jones H, Leigh FJ, Mackay I et al (2008) Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the Fertile Crescent. Mol Biol Evol 25:2211–2219https://doi.org/10.1093/molbev/msn167
Jones G, Jones H, Charles MP et al (2012) Phylogeographic analysis of barley DNA as evidence for the spread of neolithic agriculture through Europe. J Archaeol Sci 39:3230–3238https://doi.org/10.1016/j.jas.2012.05.014
Komatsuda T, Pourkheirandish M, He C et al (2007) Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA 104:1424–1429https://doi.org/10.1073/pnas.0608580104
Kota R, Varshney RK, Prasad M, Zhang H, Stein N, Graner A (2008) EST-derived single nucleotide polymorphism markers for assembling genetic and physical maps of the barley genome. Funct Integr Genomics 8:223–233. https://doi.org/10.1007/s10142-007-0060-9
Lei M, Ali M, Jiang C et al (2020) Marker-assisted selection in a global barley (Hordeum vulgare subsp. vulgare) collection revealed a unique genetic determinant of the naked barley controlled by the nud locus. Genet Resour Crop Evol 67:273–280. https://doi.org/10.1007/s10722-019-00878-3
Leino MW (2017) Spannmål: svenska lantsorter. Nordiska Museets Förlag, Stockholm
Leino MW, Hagenblad J, Edqvist J, Karlsson Strese E-M (2009) DNA preservation and utility of a historic seed collection. Seed Sci Res 19:125–135. https://doi.org/10.1017/S0960258509990055
Lister DL, Jones MK (2013) Is naked barley an eastern or a western crop? The combined evidence of archaeobotany and genetics. Veget Hist Archaeobot 22:439–446. https://doi.org/10.1007/s00334-012-0376-9
Lister DL, Thaw S, Bower MA, Jones H, Charles MP, Jones G, Smith LMJ, Howe CJ, Brown TA, Jones MK (2009) Latitudinal variation in a photoperiod response gene in european barley: insight into the dynamics of agricultural spread from ‘historic’ specimens. J Archaeol Sci 36:1092–1098. https://doi.org/10.1016/j.jas.2008.12.012
Lundequist NW (1855) Handbok i svenska landtbruket, 4th edn.Huldberg, Stockholm
Morrell PL, Clegg MT (2007) Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc Natl Acad Sci USA 104:3289–3294https://doi.org/10.1073/pnas.0611377104
Muñoz-Amatriaín M, Cuesta-Marcos A, Endelman JB et al (2014) The USDA Barley core collection: genetic diversity, population structure, and potential for genome-wide association studies. PLoS ONE 9:e94688. https://doi.org/10.1371/journal.pone.0094688
Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H et al (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3(7):e196. https://doi.org/10.1371/journal.pbio.0030196
Pasam RK, Sharma R, Walther A, Özkan H, Graner A, Kilian B (2014) Genetic diversity and population structure in a legacy collection of spring barley landraces adapted to a wide range of climates. PLoS ONE 9:e116164. https://doi.org/10.1371/journal.pone.0116164
Poets AM, Fang Z, Clegg MT, Morrell PL (2015) Barley landraces are characterized by geographically heterogeneous genomic origins. Genome Biol 16:173. https://doi.org/10.1186/s13059-015-0712-3
Pourkheirandish M, Hensel G, Kilian B et al (2015) Evolution of the grain dispersal system in barley. Cell 162:527–539. https://doi.org/10.1016/j.cell.2015.07.002
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1093/genetics/155.2.945
R Core Team (2020) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138. https://doi.org/10.1046/j.1471-8286.2003.00566.x
Saisho D, Purugganan MD (2007) Molecular phylogeography of domesticated barley traces expansion of agriculture in the Old World. Genetics 177:1765–1776. https://doi.org/10.1534/genetics.107.079491
Saisho D, Pourkheirandish M, Kanamori H, Matsumoto T, Komatsuda T (2009) Allelic variation of row type gene Vrs1 in barley and implication of the functional divergence. Breed Sci 59:621–628
Takahashi R (1955) The origin and evolution of cultivated barley. Adv Genet 7:227–266. https://doi.org/10.1016/S0065-2660(08)60097-8
Taketa S, Kikuchi S, Awayama T, Yamamoto S, Ichii M, Kawasaki S (2004) Monophyletic origin of naked barley inferred from molecular analyses of a marker closely linked to the naked caryopsis gene (nud). Theor Appl Genet 108:1236–1242. https://doi.org/10.1007/s00122-003-1560-1
Taketa S, Amano S, Tsujino Y et al (2008) Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc Natl Acad Sci USA 105:4062–4067. https://doi.org/10.1073/pnas.0711034105
Vanhanen S (2019) Prehistoric cultivation and plant gathering in Finland: An archaeobotanical study. Dissertation, Helsinki University, Helsinki
Viklund K (1998) Cereals,weeds and crop processing in Iron Age Sweden: methodological and interpretive aspects of archaeobotanical evidence. Dissertation, Umeå University, Umeå
Yu S, Long H, Deng G et al (2016) A single nucleotide polymorphism of Nud converts the caryopsis type of barley (Hordeum vulgare L). Plant Mol Biol Rep 34:242–248. https://doi.org/10.1007/s11105-015-0911-9
Zeng X, Guo Y, Xu Q et al (2018) Origin and evolution of qingke barley in Tibet. Nat Commun 9:5433. https://doi.org/10.1038/s41467-018-07920-5
Zohary D, Hopf M, Weiss E (2012) Domestication of plants in the Old World: the origin and spread of domesticated plants in south-west Asia, Europe, and the Mediterranean Basin, 4th edn. Oxford University Press, Oxford
Acknowledgements
The authors wish to thank Ida Gustafson and Karin Karlsson for assistance with laboratory work.
Funding
Open access funding provided by Stockholm University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Communicated by K.-E. Behre.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hagenblad, J., Abbey-Lee, R., Bashford, L. et al. The introduction history of Hordeum vulgare var. nudum (naked barley) into Fennoscandia. Veget Hist Archaeobot 33, 237–245 (2024). https://doi.org/10.1007/s00334-023-00925-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00334-023-00925-1