Abstract
Objective
Test a practical realignment approach to compensate the technical variability of MR radiomic features.
Methods
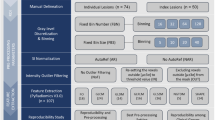
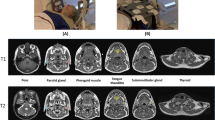
T1 phantom images acquired on 2 scanners, FLAIR and contrast-enhanced T1-weighted (CE-T1w) images of 18 brain tumor patients scanned on both 1.5-T and 3-T scanners, and 36 T2-weighted (T2w) images of prostate cancer patients scanned in one of two centers were investigated. The ComBat procedure was used for harmonizing radiomic features. Differences in statistical distributions in feature values between 1.5- and 3-T images were tested before and after harmonization. The prostate studies were used to determine the impact of harmonization to distinguish between Gleason grades (GGs).
Results
In the phantom data, 40 out of 42 radiomic feature values were significantly different between the 2 scanners before harmonization and none after. In white matter regions, the statistical distributions of features were significantly different (p < 0.05) between the 1.5- and 3-T images for 37 out of 42 features in both FLAIR and CE-T1w images. After harmonization, no statistically significant differences were observed. In brain tumors, 41 (FLAIR) or 36 (CE-T1w) out of 42 features were significantly different between the 1.5- and 3-T images without harmonization, against 1 (FLAIR) or none (CE-T1w) with harmonization. In prostate studies, 636 radiomic features were significantly different between GGs after harmonization against 461 before. The ability to distinguish between GGs using radiomic features was increased after harmonization.
Conclusion
ComBat harmonization efficiently removes inter-center technical inconsistencies in radiomic feature values and increases the sensitivity of studies using data from several scanners.
Key Points
• Radiomic feature values obtained using different MR scanners or imaging protocols can be harmonized by combining off-the-shelf image standardization and feature realignment procedures.
• Harmonized radiomic features enable one to pool data from different scanners and centers without a substantial loss of statistical power caused by intra- and inter-center variability.
• The proposed realignment method is applicable to radiomic features from different MR sequences and tumor types and does not rely on any phantom acquisition.



Similar content being viewed by others
Abbreviations
- CE-T1w:
-
Contrast-enhanced T1-weighted
- CT:
-
Computed tomography
- D1/D2:
-
Prostate cancer patient database 1/2
- hWS:
-
Hybrid white stripe
- LDA:
-
Linear discriminant analysis
- MRI:
-
Magnetic resonance imaging
- PET:
-
Positron emission tomography
- ROI:
-
Region of interest
- T2w:
-
T2-weighted
- VOI:
-
Volume of interest
- WM:
-
White matter
References
Yan J, Chu-Shern JL, Loi HY et al (2015) Impact of image reconstruction settings on texture features in 18F-FDG PET. J Nucl Med 56:1667–1673
Berenguer R, Pastor-Juan MDR, Canales-Vázquez J et al (2018) Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology 288:407–415
Goya-Outi J, Orlhac F, Calmon R et al (2018) Computation of reliable textural indices from multimodal brain MRI: suggestions based on a study of patients with diffuse intrinsic pontine glioma. Phys Med Biol 63:105003
Reuzé S, Orlhac F, Chargari C et al (2017) Prediction of cervical cancer recurrence using textural features extracted from 18F-FDG PET images acquired with different scanners. Oncotarget 8:43169–43179
Boellaard R, Delgado-Bolton R, Oyen WJG et al (2015) FDG PET/CT: EANM procedure guidelines for tumour imaging: version 2.0. Eur J Nucl Med Mol Imaging 42:328–354
Clarke LP, Nordstrom RJ, Zhang H et al (2014) The quantitative imaging network: NCI’s historical perspective and planned goals. Transl Oncol 7:1–4
Shafiq-Ul-Hassan M, Latifi K, Zhang G, Ullah G, Gillies R, Moros E (2018) Voxel size and gray level normalization of CT radiomic features in lung cancer. Sci Rep 8:10545
Mackin D, Fave X, Zhang L et al (2017) Harmonizing the pixel size in retrospective computed tomography radiomics studies. PLoS One 12:e0178524
Chatterjee A, Vallières M, Dohan A et al (2019) Creating robust predictive radiomic models for data from independent institutions using normalization. IEEE TRPMS 3:210–215
Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8:118–127
Orlhac F, Boughdad S, Philippe C et al (2018) A postreconstruction harmonization method for multicenter radiomic studies in PET. J Nucl Med 59:1321–1328
Orlhac F, Frouin F, Nioche C, Ayache N, Buvat I (2019) Validation of a method to compensate multicenter effects affecting CT radiomics. Radiology 291:53–59
Mahon RN, Ghita M, Hugo GD, Weiss E (2020) ComBat harmonization for radiomic features in independent phantom and lung cancer patient computed tomography datasets. Phys Med Biol 65:015010
Zhuge Y, Udupa JK (2009) Intensity standardization simplifies brain MR image segmentation. Comput Vis Image Underst 113:1095–1103
Ge Y, Udupa JK, Nyúl LG, Wei L, Grossman RI (2000) Numerical tissue characterization in MS via standardization of the MR image intensity scale. J Magn Reson Imaging 12:715–721
Nyúl LG, Udupa JK (1999) On standardizing the MR image intensity scale. Magn Reson Med 42:1072–1081
Shinohara RT, Sweeney EM, Goldsmith J et al (2014) Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin 6:9–19
Kickingereder P, Bonekamp D, Nowosielski M et al (2016) Radiogenomics of glioblastoma: machine learning-based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology 281:907–918
Fortin J-P, Cullen N, Sheline YI et al (2018) Harmonization of cortical thickness measurements across scanners and sites. Neuroimage 167:104–120
Lucia F, Visvikis D, Vallières M et al (2018) External validation of a combined PET and MRI radiomics model for prediction of recurrence in cervical cancer patients treated with chemoradiotherapy. Eur J Nucl Med Mol Imaging 46:864–877
Whitney HM, Li H, Ji Y, Liu P, Giger ML (2020) Harmonization of radiomic features of breast lesions across international DCE-MRI datasets. J Med Imaging (Bellingham) 7:012707
Wang H, Zhang J, Bao S et al (2020) Preoperative MRI-based radiomic machine-learning nomogram may accurately distinguish between benign and malignant soft-tissue lesions: a two-center study. J Magn Reson Imaging. https://doi.org/10.1002/jmri.27111
Zhang L-L, Huang M-Y, Li Y et al (2019) Pretreatment MRI radiomics analysis allows for reliable prediction of local recurrence in non-metastatic T4 nasopharyngeal carcinoma. EBioMedicine 42:270–280
Penzias G, Singanamalli A, Elliott R et al (2018) Identifying the morphologic basis for radiomic features in distinguishing different Gleason grades of prostate cancer on MRI: preliminary findings. PLoS One 13:e0200730
Jackson EF, Barboriak DP, Bidaut LM, Meyer CR (2009) Magnetic resonance assessment of response to therapy: tumor change measurement, truth data and error sources. Transl Oncol 2:211–215
Clark K, Vendt B, Smith K et al (2013) The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26:1045–1057
Nioche C, Orlhac F, Boughdad S et al (2018) LIFEx: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res 78:4786–4789
Zwanenburg A, Vallières M, Abdalah MA et al (2020) The Image Biomarker Standardization Initiative: standardized quantitative radiomics for high throughput image-based phenotyping. Radiology 295:328–338
Orlhac F, Soussan M, Chouahnia K, Martinod E, Buvat I (2015) 18F-FDG PET-derived textural indices reflect tissue-specific uptake pattern in non-small cell lung cancer. PLoS One 10:e0145063
Jenkinson M, Bannister P, Brady M, Smith S (2002) Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17:825–841
Tustison NJ, Avants BB, Cook PA et al (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29:1310–1320
Nyúl LG, Udupa JK, Zhang X (2000) New variants of a method of MRI scale standardization. IEEE Trans Med Imaging 19:143–150
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodology 57:289–300
Qu L, Wang S, Yap P-T, Shen D (2019) Wavelet-based semi-supervised adversarial learning for synthesizing realistic 7T from 3T MRI. Med Image Comput Comput Assist Interv 11767:786–794
Zhong J, Wang Y, Li J et al (2020) Inter-site harmonization based on dual generative adversarial networks for diffusion tensor imaging: application to neonatal white matter development. Biomed Eng Online 19:4
Modanwal G, Vellal A, Buda M, Mazurowski MA (2020) MRI image harmonization using cycle-consistent generative adversarial network. Medical Imaging 2020: Computer-Aided Diagnosis. https://doi.org/10.1117/12.2551301.
Acknowledgments
We thank Dr. J-P Fortin for making his ComBat function available to the scientific community.
Funding
This study has received funding by the “Lidex-PIM” project (Paris-Saclay University, ANR-11-IDEX-0003-02).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Fanny Orlhac.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors has significant statistical expertise (Fanny Orlhac).
Informed consent
Written informed consent was waived by the institutional review board.
Ethical approval
Institutional review board approval was obtained.
Study subjects or cohorts overlap
Some study subjects or cohorts have been previously reported in the study of Penzias et al [24].
Methodology
• retrospective
• experimental
• multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Orlhac, F., Lecler, A., Savatovski, J. et al. How can we combat multicenter variability in MR radiomics? Validation of a correction procedure. Eur Radiol 31, 2272–2280 (2021). https://doi.org/10.1007/s00330-020-07284-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07284-9