Abstract
Objectives
To propose and evaluate a method to reduce variability in emphysema quantification among different computed tomography (CT) reconstructions by normalizing CT data reconstructed with varying kernels.
Methods
We included 369 subjects from the COPDGene study. For each subject, spirometry and a chest CT reconstructed with two kernels were obtained using two different scanners. Normalization was performed by frequency band decomposition with hierarchical unsharp masking to standardize the energy in each band to a reference value. Emphysema scores (ES), the percentage of lung voxels below -950 HU, were computed before and after normalization. Bland-Altman analysis and correlation between ES and spirometry before and after normalization were compared. Two mixed cohorts, containing data from all scanners and kernels, were created to simulate heterogeneous acquisition parameters.
Results
The average difference in ES between kernels decreased for the scans obtained with both scanners after normalization (7.7 ± 2.7 to 0.3 ± 0.7; 7.2 ± 3.8 to -0.1 ± 0.5). Correlation coefficients between ES and FEV1, and FEV1/FVC increased significantly for the mixed cohorts.
Conclusions
Normalization of chest CT data reduces variation in emphysema quantification due to reconstruction filters and improves correlation between ES and spirometry.
Key Points
• Emphysema quantification is sensitive to the reconstruction kernel used.
• Normalization allows comparison of emphysema quantification from images reconstructed with varying kernels.
• Normalization allows comparison of emphysema quantification obtained with scanners from different manufacturers.
• Normalization improves correlation of emphysema quantification with spirometry.
• Normalization can be used to compare data from different studies and centers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chronic Obstructive Pulmonary Disease (COPD) is defined as a respiratory disease characterized mainly by chronic airflow limitation that is not fully reversible. It is a leading cause of death and chronic morbidity worldwide, and it represents a major public health problem [1]. The airflow limitation often presents as dyspnoea and is caused by airway disease (chronic bronchitis) or destruction of lung parenchyma (emphysema). Computed tomography (CT) allows visualization of pathologic changes in the lung parenchyma and classification of patients into different phenotypes according to the presence of bronchitis or emphysema [2]. CT analysis of lung attenuation is commonly used to quantify the extent of emphysema in the lungs by computing the emphysema score (ES): the percentage of voxels in the lung below a certain Hounsfield Unit (HU). ES is an accepted way of measuring the extent of emphysema and has been proven to correlate well with pulmonary function tests and pathology [3–7]. It is well known that ES values are sensitive to, among other factors, scanner type, dose, slice thickness, and reconstruction filter kernel used [8]. Previous studies have shown that the chosen filter kernel strongly affects emphysema quantification [9, 10], where sharper (higher spatial resolution) reconstruction kernels generally result in higher ES than smoother (lower spatial resolution) kernels. As a result, it is impossible to make meaningful comparisons between emphysema quantifications from scans obtained with different parameters. This is an important issue for longitudinal and multi-center studies where it may be difficult or impossible to control scan parameter settings.
The purpose of this retrospective study was to reduce the variability in emphysema quantification by developing a method to normalize scans obtained with different scanners and reconstructed with different kernels, and to evaluate its effectiveness in a cohort of COPD patients from a multi-center study. We hypothesized that normalization of chest CTs with respect to the filter kernel would reduce the variability in emphysema quantification.
Materials and methods
Patient selection
The COPDGene Study is a multi-center observational investigation designed to analyse the genetic and epidemiologic factors associated with COPD [11]. This study recruited 10,364 subjects from 21 institutions, with inclusion and exclusion criteria as described in [11]. All subjects underwent spirometry and CT imaging of the chest at full inspiration (TLC) and relaxed expiration (FRC) [11]. This research protocol has obtained institutional review board approval at every site and written informed consent was provided by all enrolees. For this work, the COPDGene study provided 366 scans from 183 subjects, obtained at one institution with two Siemens scanners, and 372 scans from 186 subjects, acquired at six institutions with three GE scanners. Characteristics of the patients included in our study are provided in Tables 1 and 2.
Imaging protocol
Whole-lung volumetric multi-detector CT was obtained with scanners from two different manufacturers: Siemens and GE. All scans were reconstructed applying filtered back projection (FBP) with two kernels: a standard one and a sharp one. For both manufacturers, CT scans were acquired at full inspiration without contrast medium at 120 KVp tube energy and 200 mAs effective dose. The reconstruction field-of-view was configured per patient to encompass the widest diameter of the lungs.
In the Siemens group, a Siemens Definition scanner (n = 122, 64 × 0.6 mm detector configuration, 1.1 pitch) or a Siemens Definition AS+ scanner (n = 61, 128 × 0.6 mm detector configuration, 1.0 pitch) were used. All images were reconstructed with two kernels: b31f (standard) and b45f (sharp), using a reconstruction field-of-view ranging from 260 to 410 mm and a 512 × 512 matrix, yielding a pixel size between 0.5 and 0.8 mm. The slice thickness used was 0.75 mm with an interval of 0.5 mm.
In the GE group, scans were taken using a GE LightSpeed 16 scanner (n = 105, 16 × 0.625 mm detector configuration, 1.375 pitch), a GE LightSpeed VCT scanner (n = 71, 16 × 0.625 mm detector configuration, 1.375 pitch) or a GE LightSpeed Pro 16 scanner (n = 10, 16 × 0.625 mm detector configuration, 1.375 pitch). Scans were reconstructed with STANDARD (standard) and BONE (sharp) kernels. All subjects underwent CT imaging with 0.625 mm slice thickness with an interval of 0.625 mm. The reconstruction field-of-view ranged from 260 to 500 mm and a 512 × 512 matrix, resulting in a pixel size between 0.5 and 1 mm.
Six virtual cohorts were constructed for analysis: four cohorts using scans reconstructed with a single kernel (b31f Siemens (n = 183), b45f Siemens (n = 183), STANDARD GE (n = 186), BONE GE (n = 186)) and two mixed cohorts. The single kernel cohorts allow direct comparison between standard and sharp kernels. They were used to illustrate the variation in emphysema scoring due to the reconstruction kernels, and to demonstrate that normalization reduces this variation while maintaining correlations with lung function parameters. The first mixed cohort contained all scans reconstructed with the standard kernel on the Siemens and GE scanners (n = 369). This mixed cohort represents a typical situation in multi-center studies in which different sites have different scanners and the most similar reconstruction settings are chosen. The second mixed cohort (n = 369) was constructed to contain scans with both standard and sharp kernels from both manufacturers: from the Siemens group, the b31f reconstruction was chosen for 92 randomly selected subjects, and the b45f reconstruction for the remaining 91 subjects. From the GE group, the STANDARD kernel was selected for 93 randomly selected subjects and the BONE kernel for the remaining 93 subjects. This mixed cohort simulates a possible situation in which scans from different studies are combined, or scans are retrospectively collected from different centers with different acquisition protocols.
Pulmonary function tests
Spirometry was performed using an EasyOne spirometer (ndd Medical Technologies, Andover, MA). The mean time between CT imaging and spirometry was 10.45 days (range 0–98).
Emphysema quantification
Quantification of emphysema was performed using CIRRUS Lung 13.08 (http://cirrus.diagnijmegen.nl, Diagnostic Image Analysis Group, Nijmegen, The Netherlands; Fraunhofer MEVIS, Bremen, Germany). As a first step, the lungs were automatically segmented using methods based on region growing and morphological operations [12]. All segmentations were visually checked and corrected when needed in CIRRUS Lung. The percentage of lung affected by emphysema was quantified in terms of ES, using -950 HU as an attenuation threshold.
To rule out possible variations in ES due to segmentation differences in data reconstructed with different filter kernels, we segmented the lungs in the image reconstructed with the standard kernel, and used this segmentation for both reconstructions.
Normalization
To reduce the variability in measured ES, the proposed method changes the appearance of data obtained with various reconstruction kernels so that it will have similar characteristics as a chosen reference reconstruction. The rationale behind the proposed normalization is the fact that filter kernels affect spatial resolution and image noise of the reconstructed data: sharper reconstruction kernels will preserve higher spatial frequencies but increase image noise [8].
The proposed normalization decomposes the scan into frequency bands and alters the energy in each band according to the average energies observed in the set of scans reconstructed with a reference kernel. This will reduce image noise to a similar level. In this study, we selected Siemens b31f as the reference kernel. For a detailed description of the algorithm, see Appendix 1.
Statistical analysis
Statistical analysis was performed using statistical software (IBM SPSS Statistics, Release 20.0; IBM Corp, Armonk, NY). Medians and inter-quartile ranges were computed for the non-normally distributed ES. Differences in ES for two reconstructions of the same scan were computed as ES in scans reconstructed with the sharp kernel minus ES in scans reconstructed with the standard kernel. Mean and standard deviations were computed for the normally distributed differences. The Bland-Altman approach [13] was used for analysis, with 95 % confidence intervals as limits of agreement.
Spearman correlation coefficients were calculated to evaluate the correlation between ES and forced expiratory volume in 1 second (FEV1) and FEV1-to-forced vital capacity (FVC) ratio. The statistical significance of the difference between correlation values was evaluated using the statistical test described by Steiger [14], using the R statistical analysis package (R Foundation for Statistical Computing, Vienna, Austria (URL http://www.R-project.org/).
Results
ES was successfully computed in all subjects both before and after normalization. Illustrations are provided in Figs. 1 and 2.
Example sections of a scan reconstructed with two kernels in the Siemens scanner with an emphysema overlay highlighting voxels below -950HU. The upper row shows the original sections reconstructed with (A) b31f and (B) b45f kernels. The lower row shows the same sections after normalization with (C) b31f and (D) b45f kernels. The ES obtained for the (A) b31f original scan was 9.5 %, for the (B) b45f original, ES was 21.2 %, whereas for the (C) b31f normalized, ES was 10.1 % and for the (D) b45f normalized, ES was 10.9 %
Example sections of a scan reconstructed with two kernels in the GE scanner with an emphysema overlay highlighting voxels below -950HU. The upper row shows the original sections reconstructed with (A) STANDARD and (B) BONE kernels. The lower row shows the same sections after normalization with (C) STANDARD and (D) BONE kernels. The ES obtained for the (A) STANDARD original scan was 11.8 %, for the (B) BONE original, ES was 28.0 %, whereas for the (C) STANDARD normalized, ES was 7.7 % and for the (D) BONE normalized, ES was 7.0 %
Single kernel groups: standard versus sharp kernels
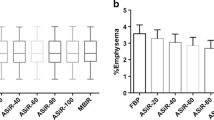
For the Siemens group, median ES were 11.6 (IQR 3.4, 23.6; range 0.1, 58.6) for data reconstructed with b31f and 21.8 (IQR 11.6, 31.0; range 0.9, 61.1) for the data reconstructed with b45f. The median ES after normalization were 16.4 (IQR 3.8, 25.5; range 0.0, 61.1) for data reconstructed with b31f and 15.4 (IQR 3.7, 25.6; range 0.0, 60.6) for data reconstructed with b45f. The average difference between ES for data reconstructed with b31f and b45f decreased from 7.7 ± 2.7 (range 0.9, 14.8) before normalization to 0.3 ± 0.7 (range -1.3, 2.4) after normalization. Figure 3 provides the Bland-Altman plots of the ES measured for the two kernels before and after normalization. Limits of agreement between ES derived from data reconstructed with b31f and b45f kernels were 2.4 % to 12.9 % before normalization. This improved to -1.0 % to 1.5 % after normalization.
In the GE group, median ES were 1.8 (IQR 0.7, 4.7; range 0.2, 51.6) and 9.6 (IQR 6.3, 16.33; range 0.8, 48.6) for scans reconstructed with the STANDARD and BONE filter kernels, respectively. After normalization, these values were 0.5 (IQR 0.2, 1.9; range 0, 52.6) for scans reconstructed with the STANDARD kernel and 0.4 (IQR 0.1, 1.7; range 0.0, 50.9) for scans reconstructed with the BONE kernel. Bland-Altman plots of the ES obtained before and after normalization are shown in Fig. 4. The average difference between ES for scans reconstructed with STANDARD and BONE kernels before normalization were 7.2 ± 3.8 (range -3.0, 17.2) and -0.1 ± 0.5 (range -3.7, 0.9) after normalization. Limits of agreement were 0.2 % to 14.6 % before normalization and -1.1 % to 0.82 % after normalization.
Table 3 summarizes the correlation of ES with spirometry measurements. ES were obtained before and after normalization for all kernels. ES showed a significant correlation with FEV1 and FEV1/FVC ratio in all four cohorts. After normalization, all correlations between emphysema quantification and lung function parameters slightly improved.
Mixed groups
For both mixed cohorts, emphysema quantification showed a significant correlation with spirometry measurements both before and after normalization (Table 3). In the mixed cohorts, the improvement of the correlations with lung function parameters was statistically significant (p < 0.05). After normalization, the correlations were the same for both mixed cohorts.
Discussion
The filter kernel used for image reconstruction has a known influence on emphysema quantification on chest CT images [9, 10], where sharper kernels lead to higher ES than smoother kernels. These effects may have implications in longitudinal or multi-center studies in which different reconstruction parameters are used. From Figs. 1 and 2, it can be observed that changing to a sharper kernel between visits could lead to false interpretations of disease progression in a longitudinal study where reconstruction settings may vary over time; for example, if scanner models and imaging software are replaced by new models. Furthermore, in the case of a multi-center study such as COPDGene, in which CT image data is obtained in various centers using several reconstruction algorithms and scanner manufacturers, these effects hamper the possibility to compare data obtained from different locations. Even though standardized imaging protocols are often used, exactly corresponding settings do not exist between manufacturers. The method presented in this paper reduces the variability in emphysema measurements due to the use of different reconstruction kernels and scanner manufacturers. The method therefore improves comparison of CT image data in settings in which reconstruction parameters are not homogeneous and can no longer be changed retrospectively. Figures 1 and 2 provide an illustration of the normalization effect in the CT data and in the emphysema mask. It can be observed that, after normalization, not only the ES values, but also the emphysema pattern, become much more similar.
The proposed normalization does not require knowledge of the proprietary reconstruction algorithms used. A set of energy coefficients is obtained by analyzing the energy in different frequency bands in a set of scans reconstructed with the desired reference kernel. These energy coefficients are then applied to the current scan to create a new normalized reconstruction. Thus, for every new scan received, regardless of the kernel or manufacturer used to reconstruct it, the same coefficients are used to create the normalized image. This makes the method very flexible, since there is no need to recompute the energy coefficients or have any knowledge of the filter kernel. In this work, we used the Siemens b31f kernel as the reference one, and we applied the energy coefficients computed in the reference to all the scans, independent of the kernel or manufacturer used to create the CT image.
Schaller et al. presented a technique to simulate smoother kernels by using a Gaussian filter to approximate the ratio between the smooth (desired) and sharp (original) kernel [15]. This method needs information about the kernel function, which is usually not publicly available. Ohkubo et al. presented an image filtering technique to create the image for a desired kernel using an existing CT reconstructed with a different kernel [16]. This image filtering uses the modulation transfer functions (MTFs) of the current kernel and the desired one. Contrary to the proposed method, the technique of Ohkubo et al. requires prior knowledge of the MTF of each kernel that may be used. The effect of the methods proposed by Schaller et al. and Ohkubo et al. on emphysema quantification has not been assessed. Bartel et al. proposed a method for equating emphysema measurements using a mathematical model to convert values between different kernels, requiring the construction of a model for every kernel separately [17]. The technique calculates an equivalent ES, but the effect of the model cannot be visually observed in the scan. Furthermore, all these methods need to be re-calibrated for every new desired kernel.
In order to ensure the validity of the normalized ES measurements, correlation coefficients between ES and lung function parameters were calculated. As shown in Table 3, for the single kernel cohorts, there are no significant differences before and after normalization, indicating that the normalization does not change the correlations with lung function parameters as expected. The correlations after normalization are slightly higher for each cohort. However, the Bland-Altman plots in Figs. 3 and 4 show that the difference in emphysema quantification between the different reconstructions of the same scans is greatly reduced by the normalization.
The first mixed cohort was obtained from a multi-center study in which standardized imaging protocols were well controlled by selecting the most similar reconstruction kernels between vendors. Even in this well-controlled setting, there is variation in emphysema measurements, because exactly corresponding acquisition parameters do not exist across vendors. There was a significant correlation between emphysema quantification and lung function parameters. However, applying the normalization significantly improved this correlation. This suggests that the use of the normalization method presented is beneficial in multi-center studies, even when the acquisition parameters are well controlled.
The second mixed cohort was constructed to simulate a less well-controlled study, e.g., when different studies with different acquisition protocols are retrospectively combined. As expected, the correlation with lung function parameters in this cohort was lower than in the well-controlled setting. Applying the normalization to this cohort not only significantly improved this correlation, but it resulted in correlation coefficients similar to those of the controlled mixed cohort. This allowed us to compare emphysema quantifications obtained from data acquired with different parameters.
From the results in Table 3, it can be observed that the correlations between emphysema quantification and spirometry are higher for the mixed cohorts than for each individual cohort. This can be explained by the difference in emphysema severity in both cohorts; the Siemens cohort contains mostly subjects with mild to severe COPD, while the GE group includes mostly subjects with no or mild COPD. Combining the cohorts includes a more complete range of COPD and emphysema severity, and therefore increases the correlation with lung function parameters.
Although the study provides promising results, it also has limitations. We validated emphysema measurements against lung function parameters, but not against histopathological references. The latter would involve subjects undergoing lobe or lung resection, which is not a feasible option in our study. Therefore, since CT quantified emphysema has been shown to correlate both with histopathological reference standards [3–5, 7] and pulmonary function testing [6, 7], we chose to compare emphysema measurements with lung function parameters.
In the current study, the normalization was evaluated in scans reconstructed with two kernels from two vendors. However, these results suggest that this idea can be extended to more reconstruction kernels from the same scanner models, and can be applied to kernels from other scanner manufacturers. This normalization method should be evaluated in a future study including data reconstructed with different kernels and scanner manufacturers.
Furthermore, in this study we have analysed only filtered back projection algorithms, but ES values have been shown to vary, depending upon the choice of filtered back projection or iterative reconstruction algorithms [18].
In conclusion, the lack of standards that guarantee the validity of CT measurements performed with different technical parameters makes it difficult to compare data obtained with different reconstruction settings. The proposed method may be a feasible solution to overcome this issue. It requires no prior knowledge about the filter kernels used, and allows one to obtain more reliable results that are independent of the reconstruction parameters chosen. Normalization of chest CT data reduces variation in emphysema quantification due to different reconstruction filters and scanner manufacturers, and improves correlation of emphysema quantifications and spirometry in data obtained with varying reconstruction settings.
Abbreviations
- COPD:
-
Chronic Obstructive Pulmonary Disease
- CT:
-
Computed Tomography
- ES:
-
Emphysema Score
- FBP:
-
Filtered Back Projection
- HU:
-
Hounsfield Unit
- TLC:
-
Total Lung Capacity
- FRC:
-
Functional Residual Capacity
- FEV1 :
-
Forced Expiratory Volume in 1 second
- FVC:
-
Forced Vital Capacity
- IQR:
-
Interquartile Range
- MTF:
-
Modulation Transfer Functions
References
Rabe KF, Hurd S, Anzueto A et al (2007) Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: Gold executive summary. Am J Respir Crit Care Med 176:532–555
Hoffman EA, Simon BA, McLennan G (2006) State of the art. A structural and functional assessment of the lung via multidetector-row computed tomography: phenotyping chronic obstructive pulmonary disease. Proc Am Thorac Soc 3:519–532
Müller NL, Staples CA, Miller RR, Abboud RT (1988) Density mask. An objective method to quantitate emphysema using computed tomography. Chest 94:782–787
Bankier AA, De Maertelaer V, Keyzer C, Gevenois PA (1999) Pulmonary emphysema: subjective visual grading versus objective quantification with macroscopic morphometry and thin-section CT densitometry. Radiology 211:851–858
Stolk J, Stoel BC (2011) Lung densitometry to assess progression of emphysema in chronic obstructive pulmonary disease: time to apply in the clinic? Am J Respir Crit Care Med 183:1578–1580
Camiciottoli G, Bartolucci M, Maluccio NM et al (2006) Spirometrically gated high-resolution CT findings in COPD: lung attenuation vs lung function and dyspnea severity. Chest 129:558–564
Madani A, van Muylem A, de Maertelaer V, Zanen J, Gevenois PA (2008) Pulmonary emphysema: size distribution of emphysematous spaces on multidetector CT images–comparison with macroscopic and microscopic morphometry. Radiology 248:1036–1041
Mets OM, de Jong PA, van Ginneken B, Gietema HA, Lammers JWJ (2012) Quantitative computed tomography in COPD: Possibilities and limitations. Lung 190:133–145
Gierada DS, Bierhals AJ, Choong CK et al (2010) Effects of CT section thickness and reconstruction kernel on emphysema quantification relationship to the magnitude of the CT emphysema index. Acad Radiol 17:146–156
Boedeker KL, McNitt-Gray MF, Rogers SR et al (2004) Emphysema: Effect of reconstruction algorithm on CT imaging measures. Radiology 232:295–301
Regan EA, Hokanson JE, Murphy JR et al (2010) Genetic epidemiology of COPD (COPDGene) study design. COPD 7:32–43
van Rikxoort EM, de Hoop B, Viergever MA, Prokop M, van Ginneken B (2009) Automatic lung segmentation from thoracic computed tomography scans using a hybrid approach with error detection. Med Phys 36:2934–2947
Bland JM, Altman DG (1986) Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1:307–310
Steiger JH (1980) Test for comparing elements in a correlation matrix. Psychol Bull 87:245–252
Schaller S, Wildberger JE, Raupach R, Niethammer M, Klingenbeck-Regn K, Flohr T (2003) Spatial domain filtering for fast modification of the tradeoff between image sharpness and pixel noise in computed tomography. IEEE Trans Med Imaging 22:846–853
Ohkubo M, Wada S, Kayugawa A, Matsumoto T, Murao K (2011) Image filtering as an alternative to the application of a different reconstruction kernel in CT imaging: feasibility study in lung cancer screening. Med Phys 38:3915–3923
Bartel ST, Bierhals AJ, Pilgram TK et al (2011) Equating quantitative emphysema measurements on different CT image reconstructions. Med Phys 38:4894–4902
Choo JY, Goo JM, Lee CH, Park CM, Park SJ, Shim MS (2014) Quantitative analysis of emphysema and airway measurements according to iterative reconstruction algorithms: comparison of filtered back projection, adaptive statistical iterative reconstruction and model-based iterative reconstruction. Eur Radiol 24:799–806
Wan ES, Hokanson JE, Murphy JR et al (2011) Clinical and Radiographic Predictors of GOLD-Unclassified Smokers in the COPDGene Study. Am J Respir Crit Care Med 184:57–63
Abe K, Katsuragawa S, Sasaki Y, Yanagisawa T (1992) A fully automated adaptive unsharp masking technique in digital chest radiograph. Invest Radiol 27:64–70
Stahl M, Aach T, Dippel S (2000) Digital radiography enhancement by nonlinear multiscale processing. Med Phys 27:56–65
Acknowledgments
The scientific guarantor of this publication is Eva van Rikxoort. The authors of this manuscript declare relationships with the following companies: MeVis Medical Solutions AG, Bremen, Germany; Thirona, Nijmegen, The Netherlands. This study has received funding by scientific grants from the Dutch Organization for Scientific Research (NWO) (VENI 016.128.007, VICI 016.130.326). No complex statistical methods were necessary for this paper. Institutional Review Board approval was obtained.
Written informed consent was obtained from all subjects (patients) in this study. Some study subjects or cohorts have been previously reported. There have been a number of publications using data from the COPDGene database for different purposes. However, none of them tries to develop a method to reduce the variability in emphysema quantifications obtained from CT scans reconstructed with different filter kernels. Some examples of publications using subjects from the COPDGene study for imaging research are: 1. Sieren JP, Newell JD, Judy PF, Lynch DA, Chan KS, Guo J, Hoffman EA (2012) Reference standard and statistical model for intersite and temporal comparisons of CT attenuation in a multicenter quantitative lung study. Med Phys 39: 5757–5767. 2. Ginsburg SB, Lynch DA, Bowler RP, Schroeder JD (2012) Automated texture-based quantification of centrilobular nodularity and centrilobular emphysema in chest CT images. Acad Radiol 19:1241-1251.
Methodology: retrospective, experimental, multicenter study.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
A1. Normalization algorithm
Every scan is decomposed into a set of frequency bands based on hierarchical unsharp masking [19, 20]. Let I denote the original image, L σ the original image convoluted with a Gaussian filter at σ scale (where L 0 = I), and n the number of frequency F i bands. Frequency bands are now defined as:
Where σ i is a set of n scales. For this paper, six frequency bands were used with values σ i = {0, 1, 2, 4, 8, 16} voxels.
The normalized image I N is constructed from the frequency bands as:
The factor λ i is used to scale the energy present in each frequency band to a reference value in each scan, ensuring that all scans will have the same energy in each frequency band after normalization. Note that if all λ i equal 1, the original image is recovered. The energy of a band, e i , is expressed as the standard deviation of the HU in the lungs. To determine the target energy for each frequency band, the average energy r i in the segmented lungs in each band in a set of scans reconstructed with the reference kernel is used. To achieve a more uniform normalization, Eq. 2 is iteratively applied until each λ i deviated at most 0.05 from 1, i.e., 0.95 ≤ λ i ≤ 1.05 for 1 ≤ i ≤ 5. The coefficients r i obtained for the reference kernel were {70.34, 67.54, 60.90, 51.45, 36.14}.
A2. Results: emphysema scores versus pulmonary function tests
This section illustrates the correlation between ES values and FEV1, and ES and FEV1/FVC. Figure A1 shows the relation between ES and PFT for each scan. As observed in Table 3, the correlation for the mixed original (two kernels) cohort is higher than for any of the single scanner cohorts. By combining the single scanner cohorts into the mixed one, the range of emphysema severity is more complete, as the Siemens cohort contains mostly subjects with mild to severe COPD, while the GE group includes mostly subjects with no or mild COPD. This produces an increase of the correlation with lung function parameters.
Scatter plots showing ES values versus FEV1 (A) and FEV1/FVC (B). The black circles represent the patients in the GE STANDARD original cohort and the white diamonds represent the patients in the Siemens b31f original cohort. The mixed original cohort (two kernels) is the combination of the two mentioned cohorts
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Gallardo-Estrella, L., Lynch, D.A., Prokop, M. et al. Normalizing computed tomography data reconstructed with different filter kernels: effect on emphysema quantification. Eur Radiol 26, 478–486 (2016). https://doi.org/10.1007/s00330-015-3824-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-015-3824-y