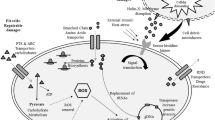
Abstract
Cryobacterium spp. are Gram-positive bacteria that inhabit diverse geographical locations, particularly extremely cold environments like the Polar Regions. However, strategies that enable them to survive in harsh Antarctic environments are not fully understood. In this study, we conducted a comparative genomic analysis of the Antarctic Cryobacterium sp. SO2 as well as other members of the Cryobacterium genus. Phylogenetic analysis revealed that strain SO2 formed a distinct cluster with the validly described species: C. adonitolivorans, C. lactosi, C. soli, C. arcticum, and C. zongtaii. Comparative analysis based on ANI and AAI indicated that strain SO2 is a novel species. Relative Synonymous Codon Usage (RSCU) of Cryobacterium species exhibits a bias towards codon ending with G/C. The genomes of all strains harbored numerous genes associated with environmental-associated stress responses, including oxidative stress response, general stress response, heat-stress response, cold-stress response, cell envelope alteration, and osmotic stress response. Strain SO2 and related strains possess genes involved in breaking down and utilization of both plant and animal carbohydrate-containing materials. KEGG annotation indicated that strain SO2 and related species shared almost the same genes for the metabolism of trehalose (TreS, partial TPS/TPP, and TreY-TreZ pathway) and glycogen (classical and non-classical pathway). The results from this work helped us to better understand the genomic characteristics of Cryobacterium spp. in terms of genomic diversity and adaptation strategies, which may have significant implications for biotechnology and climate change research.








Similar content being viewed by others
Change history
26 March 2024
A Correction to this paper has been published: https://doi.org/10.1007/s00300-024-03233-0
References
Afratis N, Gialeli C, Nikitovic D, Tsegenidis T, Karousou E, Theocharis AD, Pavão MS, Tzanakakis GN, Karamanos NK (2012) Glycosaminoglycans: key players in cancer cell biology and treatment. FEBS J 279(7):1177–1197. https://doi.org/10.1111/j.1742-4658.2012.08529.x
Alscher RG, Erturk N, Heath LS (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53(372):1331–1341
Aschenbrenner IA, Cernava T, Berg G, Grube M (2016) Understanding microbial multi-species symbioses. Front Microbiol 7:180. https://doi.org/10.3389/fmicb.2016.00180
Avonce N, Mendoza-Vargas A, Morett E, Iturriaga G (2006) Insights on the evolution of trehalose biosynthesis. BMC Evol Biol 6(1):109. https://doi.org/10.1186/1471-2148-6-109
Bagautdinov B, Ukita Y, Miyano M, Kunishima N (2008) Structure of 3-oxoacyl-(acyl-carrier protein) synthase II from Thermus C. thermophilus HB8. Acta Crystallogr Sect F Struct Biol Cryst Commun 64(Pt5):358–366. https://doi.org/10.1107/S1744309108010336
Bajerski F, Ganzert L, Mangelsdorf K, Lipski A, Wagner D (2011) Cryobacterium arcticum sp. Nov., a psychrotolerant bacterium from an Arctic soil. Int J Syst Evol Microbiol 61(8):1849–1853. https://doi.org/10.1099/ijs.0.027128-0
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37(Database issue):D233–D238. https://doi.org/10.1093/nar/gkn663
Chattopadhyay M, Jagannadham M (2001) Maintenance of membrane fluidity in Antarctic bacteria. Polar Biol 24(5):386–388. https://doi.org/10.1007/s003000100232
Colesie C, Walshaw CV, Sancho LG, Davey MP, Gray A (2023) Antarctica’s vegetation in a changing climate. WIREs Climate Change 14(1):e810. https://doi.org/10.1002/wcc.810
Cui S, Hu M, Sun Y, Mao B, Zhang Q, Zhao J, Tang X, Zhang H (2022) Effect of trehalose and lactose treatments on the freeze-drying resistance of lactic acid bacteria in high-density culture. Microorganisms 11(1):48. https://doi.org/10.3390/microorganisms11010048
Dieser M, Greenwood M, Foreman CM (2010) Carotenoid pigmentation in Antarctic heterotrophic bacteria as a strategy to withstand environmental stresses. Arct Antarct Alp Res 42(4):396–405. https://doi.org/10.1657/1938-4246-42.4.396
Eddy SR (2011) Accelerated profile HMM searches. PloS Comput Biol 7(1):e1002195. https://doi.org/10.1371/journal.pcbi.1002195
Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO (2015) Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3:e1319. https://doi.org/10.7717/peerj.1319
Fong NJ, Burgess ML, Barrow KD, Glenn DR (2001) Carotenoid accumulation in the psychrotrophic bacterium Arthrobacter agilis in response to thermal and salt stress. Appl Microbiol Biotechnol 56(5–6):750–756. https://doi.org/10.1007/s002530100739
Gao H, Wang Y, Liu X, Yan T, Wu L, Alm E, Arkin A, Thompson DK, Zhou J (2004) Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J Bacteriol 186(22):7796–7803. https://doi.org/10.1128/JB.186.22.7796-7803.2004
Garcia-Rubio R, de Oliveira HC, Rivera J, Trevijano-Contador N (2020) The fungal cell wall: Candida, Cryptococcus, and Aspergillus species. Front Microbiol 10:2993. https://doi.org/10.3389/fmicb.2019.02993
Goordial J, Raymond-Bouchard I, Zolotarov Y et al (2015) Cold adaptive traits revealed by comparative genomic analysis of the eurypsychrophile Rhodococcus sp. JG3 isolated from high elevation McMurdo Dry Valley permafrost. Antarctica. FEMS Microbiol Ecol 92:fiv154. https://doi.org/10.1093/femsec/fiv154
Grisaro V, Sharon N, Barkai-Golan R (1968) The chemical composition of the cell walls of Penicillium digitatum Sacc. And Penicillium italicum Whem. J Gen Microbiol 51(1):145–150. https://doi.org/10.1099/00221287-51-1-145
Guo Q-Q, Zhang W-B, Zhang C et al (2019) Characterization of 3-oxacyl-acyl carrier protein reductase homolog genes in Pseudomonas aeruginosa PAO1. Front Microbiol 10:1028. https://doi.org/10.3389/fmicb.2019.01028
Guo R, Liu T, Guo C, Chen G, Fan J, Zhang Q (2022) Carotenoid biosynthesis is associated with low-temperature adaptation in Rhodosporidium kratochvilovae. BMC Microbiol 22(1):319. https://doi.org/10.1186/s12866-022-02728-2
Han L, Lobo S, Reynolds KA (1998) Characterization of beta-ketoacyl-acyl carrier protein synthase III from Streptomyces glaucescens and its role in initiation of fatty acid biosynthesis. J Bacteriol 180(17):4481–4486. https://doi.org/10.1128/JB.180.17.4481-4486.1998
Herdeiro RS, Pereira MD, Panek AD, Eleutherio ECA (2006) Trehalose protects Saccharomyces cerevisiae from lipid peroxidation during oxidative stress. Biochimica et Biophysica Acta (BBA) – General Subjects 1760(3):340–346. https://doi.org/10.1016/j.bbagen.2006.01.010
Hoffmann T, Bremer E (2011) Protection of Bacillus subtilis against cold stress via compatible-solute acquisition. J Bacteriol 193(7):1552–1562. https://doi.org/10.1128/JB.01319-10
Hubloher JJ, Zeidler S, Lamosa P, Santos H, Averhoff B, Müller V (2020) Trehalose-6-phosphate-mediated phenotypic change in Acinetobacter baumannii. Environ Microbiol 22(12):5156–5166. https://doi.org/10.1111/1462-2920.15148
Huerta-Cepas J, Szklarczyk D, Forslund K et al (2016) eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44(1):D286–D293. https://doi.org/10.1093/nar/gkv1248
Huerta-Cepas J, Forslund K, Coelho LP et al (2017) Fast Genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol Biol Evol 34(8):2115–2122. https://doi.org/10.1093/molbev/msx148
Hwang G, Kim S, Cho J-Y et al (2019) Trehalose-6-phosphate signaling regulates thermoresponsive hypocotyl growth in Arabidopsis thaliana. EMBO Rep 20(10):e47828. https://doi.org/10.15252/embr.201947828
Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S (2018) High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9(1):5114. https://doi.org/10.1038/s41467-018-07641-9
Jiang M, Pang X, Chen H, Chen D (2022) Ecological integrity evaluation along the Antarctic coast: a case study on the Fildes Peninsula. Cont Shelf Res 242:104747. https://doi.org/10.1016/j.csr.2022.104747
Jin M, Xiao A, Zhu L, Zhang Z, Huang H, Jiang L (2019) The diversity and commonalities of the radiation-resistance mechanisms of Deinococcus and its up-to-date applications. AMB Express 9(1):138. https://doi.org/10.1186/s13568-019-0862-x
Kandror O, DeLeon A, Goldberg AL (2002) Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc Natl Acad Sci 99(15):9727–9732. https://doi.org/10.1073/pnas.142314099
Kanehisa M, Sato Y, Morishima K (2016) BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428(4):726–731. https://doi.org/10.1016/j.jmb.2015.11.006
Kedzierska S, Matuszewska E (2001) The effect of co-overproduction of DnaK/DnaJ/GrpE and ClpB proteins on the removal of heat-aggregated proteins from Escherichia coliΔclpB mutant cells-new insight into the role of Hsp70 in a functional cooperation with Hsp100. FEMS Microbiol Lett 204(2):355–360. https://doi.org/10.1111/j.1574-6968.2001.tb10910.x
Kempf B, Bremer E (1995) OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. JBC 270(28):16701–16713. https://doi.org/10.1074/jbc.270.28.16701
Keto-Timonen R, Hietala N, Palonen E et al (2016) Cold shock proteins: a minireview with special emphasis on Csp-family of Enteropathogenic Yersinia. Front Microbiol 7:1151. https://doi.org/10.3389/fmicb.2016.01151
Kim D, Park S, Chun J (2021) Introducing EzAAI: a pipeline for high throughput calculations of prokaryotic average amino acid identity. J Microbiol 59(5):476–480. https://doi.org/10.1007/s12275-021-1154-0
Kolde R (2019) pheatmap: Pretty Heatmaps (R package version 1.0.12). https://CRAN.R-project.org/package=pheatmap
Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU (1992) Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356(6371):683–689. https://doi.org/10.1038/356683a0
Lee I, Ouk Kim Y, Park S-C, Chun J (2016) OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66(2):1100–1103. https://doi.org/10.1099/ijsem.0.000760
Lee A, Bae E, Park J, Choi K-H, Cha J (2021) Identification of the genes related to the glycogen metabolism in hyperthermophilic Archaeon. Front Microbiol 12:661053. https://doi.org/10.3389/fmicb.2021.661053
Li P, Harding SE, Liu Z (2001) Cyanobacterial exopolysaccharides: their nature and potential biotechnological applications. Biotechnol Genet Eng Rev 18(1):375–404. https://doi.org/10.1080/02648725.2001.10648020
Liu Q, Yang L-L, Xin Y-H (2023) Diversity of the genus Cryobacterium and proposal of 19 novel species isolated from glaciers. Front Microbiol 14:1115168. https://doi.org/10.3389/fmicb.2023.1115168
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42(D1):D490–D495. https://doi.org/10.1093/nar/gkt1178
Lu J, Holmgren A (2014) The thioredoxin antioxidant system. Free Radic Biol Med 66:75–87. https://doi.org/10.1016/j.freeradbiomed.2013.07.036
Luo D, Wu Z, Bai Q, Zhang Y, Huang M, Huang Y, Li X (2023) Universal stress proteins: from gene to function. Int J Mol Sci 24(5):4725. https://doi.org/10.3390/ijms24054725
Maleki F, Khosravi A, Nasser A, Taghinejad H, Azizian M (2016) Bacterial heat shock protein activity. J Clin Diag Res 10(3):BE01–BE03. https://doi.org/10.7860/JCDR/2016/14568.7444
Minh BQ, Schmidt HA, Chernomor O et al (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37(5):1530–1534. https://doi.org/10.1093/molbev/msaa015
Nakaminami K, Karlson DT, Imai R (2006) Functional conservation of cold shock domains in bacteria and higher plants. Proc Natl Acad Sci 103(26):10122–10127. https://doi.org/10.1073/pnas.0603168103
Nakamura A, Furuta H, Maeda H, Nagamatsu Y, Yoshimoto A (2001) Analysis of structural components and molecular construction of soybean soluble polysaccharides by stepwise enzymatic degradation. Biosci Biotechnol Biochem 65(10):2249–2258. https://doi.org/10.1271/bbb.65.2249
Oh HN, Park D, Seong HJ, Kim D, Sul WJ (2019) Antarctic tundra soil metagenome as useful natural resources of cold-active lignocelluolytic enzymes. J Microbiol 57(10):865–873. https://doi.org/10.1007/s12275-019-9217-1
Oren A, Garrity GM (2021) Valid publication of the names of forty-two phyla of prokaryotes. Int J Syst Evol Microbiol 71(10):005056. https://doi.org/10.1099/ijsem.0.005056
Park M-J, Kim YJ, Park M, Yu J, Namirimu T, Roh Y-R, Kwon KK (2022) establishment of genome based criteria for classification of the family Desulfovibrionaceae and proposal of two novel genera Alkalidesulfovibrio gen. nov. and Salidesulfovibrio gen. nov. Front Microbiol 13:738205. https://doi.org/10.3389/fmicb.2022.738205
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25(7):1043–1055. https://doi.org/10.1101/gr.186072.114
Paulin KA, Cortez D, Eichman BF (2022) The SOS response-associated peptidase (SRAP) domain of YedK catalyzes ring opening of abasic sites and reversal of its DNA–protein cross-link. J Biol Chem 298(9):102307. https://doi.org/10.1016/j.jbc.2022.102307
Perkins A, Nelson KJ, Parsonage D, Poole LB, Karplus PA (2015) Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem Sci 40(8):435–445. https://doi.org/10.1016/j.tibs.2015.05.001
Přerovská T, Henke S, Bleha R et al (2021) Arabinogalactan-like glycoproteins from Ulva lactuca (Chlorophyta) show unique features compared to land plants AGPs. J Phycol 57:619–635. https://doi.org/10.1111/jpy.13121
Rath H, Reder A, Hoffmann T et al (2020) Management of osmoprotectant uptake hierarchy in Bacillus subtilis via a SigB-Dependent Antisense RNA. Front Microbiol 11:622. https://doi.org/10.3389/fmicb.2020.00622
Reddy GSN, Pradhan S, Manorama R, Shivaji S (2010) Cryobacterium roopkundense sp. Nov., a psychrophilic bacterium isolated from glacial soil. Int J Syst Evol Microbiol 60(4):866–870. https://doi.org/10.1099/ijs.0.011775-0
Roberts AW, Roberts EM, Haigler CH (2012) Moss cell walls: structure and biosynthesis. Front Plant Sci 3:166. https://doi.org/10.3389/fpls.2012.00166
Roig-Oliver M, Douthe C, Bota J, Flexas J (2021) Cell wall thickness and composition are related to photosynthesis in Antarctic mosses. Physiol Plant 173(4):1914–1925. https://doi.org/10.1111/ppl.13533
Seel W, Baust D, Sons D et al (2020) Carotenoids are used as regulators for membrane fluidity by Staphylococcus xylosus. Sci Rep 10(1):330. https://doi.org/10.1038/s41598-019-57006-5
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068–2069. https://doi.org/10.1093/bioinformatics/btu153
Segal G, Ron EZ (1998) Regulation of heat-shock response in bacteria. Ann N Y Acad Sci 851:147–151. https://doi.org/10.1111/j.1749-6632.1998.tb08988.x
Seibold GM, Eikmanns BJ (2007) The glgX gene product of Corynebacterium glutamicum is required for glycogen degradation and for fast adaptation to hyperosmotic stress. Microbiology (reading, England) 153(Pt7):2212–2220. https://doi.org/10.1099/mic.0.2006/005181-0
Sekar K, Linker SM, Nguyen J, Grünhagen A, Stocker R, Sauer U (2020) Bacterial glycogen provides short-term benefits in changing environments. Appl Environ Microbiol 86(9):e00049. https://doi.org/10.1128/AEM.00049-20
Selye H (1936) A syndrome produced by diverse nocuous agents. Nature 138(3479):32. https://doi.org/10.1038/138032a0
Selye H (1950) Stress and the general adaptation syndrome. BMJ 1(4667):1383–1392. https://doi.org/10.1136/bmj.1.4667.1383
Steenwyk JL, Buida TJ, Gonçalves C et al (2022) BioKIT: a versatile toolkit for processing and analyzing diverse types of sequence data. Genetics 221(3):iyac079. https://doi.org/10.1093/genetics/iyac079
Styczynski M, Rogowska A, Gieczewska K, Garstka M, Szakiel A, Dziewit L (2020) Genome-based insights into the production of carotenoids by Antarctic bacteria, Planococcus sp. ANT_H30 and Rhodococcus sp. ANT_H53B. Molecules 25(19):4357. https://doi.org/10.3390/molecules25194357
Suzuki K-I, Sasaki J, Uramoto M et al (1997) Cryobacterium psychrophilum gen. nov., sp. Nov., nom. Rev., comb. Nov., an obligately psychrophilic Actinomycete to accommodate “Curtobacterium psychrophilum” Inoue and Komagata 1976. Int J Syst Bacteriol 47(2):474–478. https://doi.org/10.1099/00207713-47-2-474
Teoh CP, Wong CMVL, Lee DJH, González-Aravena M, Najimudin N, Lee PC, Cheah YK (2018) Genome sequences of two cold-adapted Cryobacterium spp. SO1 and SO2 from Fildes Peninsula, Antarctica. Curr Sci 115(9):1706–1708. https://doi.org/10.18520/cs/v115/i9/1706-1708
Teoh CP, Lavin P, Lee DJH, González-Aravena M, Najimudin N, Lee PC, Cheah YK, Wong CMVL (2021) Genomics and transcriptomics analyses provide insights into the cold adaptation strategies of an Antarctic bacterium, Cryobacterium sp. SO1. Polar Biol 44(7):1305–1319. https://doi.org/10.1007/s00300-021-02883-8
Teoh CP, Lavin P, González-Aravena M, Wong CMVL (2023) genomic analysis of a novel antarctic bacterium, Cyrobacterium sp. SO2 provides insights into its genomic potential for production of antimicrobial compounds. Sains Malaysiana 52(7):1955–1966. https://doi.org/10.17576/jsm-2023-5207-05
Thompson CC, Chimetto L, Edwards RA, Swings J, Stackebrandt E, Thompson FL (2013) Microbial genomic taxonomy. BMC Genomics 14(1):913. https://doi.org/10.1186/1471-2164-14-913
Vance TDR, Bayer-Giraldi M, Davies PL, Mangiagalli M (2019) Ice-binding proteins and the ‘domain of unknown function’ 3494 family. FEBS J 286(5):855–873. https://doi.org/10.1111/febs.14764
Volker U, Engelmann S, Maul B et al (1994) Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology (n y) 140:741–752. https://doi.org/10.1099/00221287-140-4-741
Wachananawat B, Kuroha T, Takenaka Y et al (2020) Diversity of pectin rhamnogalacturonan I rhamnosyltransferases in glycosyltransferase family 106. Front Plant Sci 11:997. https://doi.org/10.3389/fpls.2020.00997
Wang L, Wise MJ (2019) An updated view on bacterial glycogen structure. Microbiol Aust. https://doi.org/10.1071/MA19056
Waterman MJ, Bramley-Alves J, Miller RE, Keller PA, Robinson SA (2018) Photoprotection enhanced by red cell wall pigments in three East Antarctic mosses. Biol Res 51(1):49. https://doi.org/10.1186/s40659-018-0196-1
Winder JC, Boulton W, Salamov A et al (2023) Genetic and structural diversity of prokaryotic ice-binding proteins from the Central Arctic Ocean. Genes (basel) 14(2):363. https://doi.org/10.3390/genes14020363
Wolffe AP (1995) The cold-shock response in bacteria. Sci Prog 78(Pt 4):301–310
Zhang D-C, Wang H-X, Cui H-L et al (2007) Cryobacterium psychrotolerans sp. Nov., a novel psychrotolerant bacterium isolated from the China No. 1 glacier. Int J Syst Evol Microbiol 57(4):866–869. https://doi.org/10.1099/ijs.0.64750-0
Zhang X-F, Shen W, Gurunathan S (2016) Biologically synthesized gold nanoparticles ameliorate cold and heat stress-induced oxidative stress in Escherichia coli. Molecules 21(6):731. https://doi.org/10.3390/molecules21060731
Zhang D, Hu P, Liu T, Wang J, Jiang S, Xu Q, Chen L (2018a) GC bias lead to increased small amino acids and random coils of proteins in cold-water fishes. BMC Genomics 19:315. https://doi.org/10.1186/s12864-018-4684-z
Zhang H, Yohe T, Huang L et al (2018b) dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46(W1):W95–W101. https://doi.org/10.1093/nar/gky418
Zheng J, Ge Q, Yan Y, Zhang X, Huang L, Yin Y (2023) dbCAN3: automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res 51(W1):W115–W121. https://doi.org/10.1093/nar/gkad328
Zúñiga-González P, Zúñiga GE, Pizarro M, Casanova-Katny A (2016) Soluble carbohydrate content variation in Sanionia uncinata and Polytrichastrum alpinum, two Antarctic mosses with contrasting desiccation capacities. Biol Res 49:6. https://doi.org/10.1186/s40659-015-0058-z
Acknowledgements
The research work described in this paper was supported by Funding from the Ministry of Science, Technology, and Innovation (MOSTI), Malaysia under the Antarctic Flagship Project (FP1213E36).
Author information
Authors and Affiliations
Contributions
CMVLW conceived the research. TCP designed and conducted experiments. TCP performed the data analyses. GMA and PL facilitated the collection of samples for the isolation of bacteria. CMVLW and TCP wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: There is a typo in species name and they have been corrected in the original version.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Teoh, C.P., González‑Aravena, M., Lavin, P. et al. Cold adaptation and response genes of Antarctic Cryobacterium sp. SO2 from the Fildes Peninsula, King George Island. Polar Biol 47, 135–156 (2024). https://doi.org/10.1007/s00300-023-03213-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-023-03213-w