Abstract
Key message
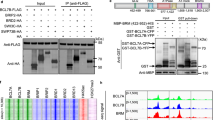
Chromatin remodeling ATPases OsSYD and OsBRM are involved in shoot establishment, and both affect OSH gene transcription. OsSYD protein interacts with RFL, but OsBRM does not.
Abstract
In plants, SPLAYED (SYD) and BRAHMA (BRM) encode chromatin remodeling ATPases that use the energy derived from ATP hydrolysis to restructure nucleosomes and render certain genomic regions available to transcription factors. However, the function of SYD and BRM on rice growth and development is unknown. Here, we constructed ossyd and osbrm mutants using CRISPR/Cas9 technology and analyzed the effects of mutations on rice embryo development. We discovered that the ossyd and osbrm mutants exhibited severe defects during embryonic development, whereas endosperm development was normal. These results indicated that the development of the embryo and endosperm is independent of each other. Consequently, the ossyd- and osbrm-null mutants did not germinate due to the abnormal embryos. Furthermore, we observed the embryos of ossyd- and osbrm-null mutants, and they indeed had distinct differentiation defects in shoot establishment, acquired during embryogenesis. To verify the function of OsSYD and OsBRM in embryogenesis, we measured the transcript levels of marker genes at different stages. Compared with wild type, the expression levels of multiple OSH genes were significantly reduced in the mutants, which was consistent with the defective shoot establishment phenotypes. The interaction between SYD and RICE FLORICAULA/LFY (RFL) was revealed using a yeast two-hybrid screening system, suggesting that the interaction between the LFY homolog and chromatin remodeling ATPases is ubiquitous in plants. Collectively, our findings provide the basis for elucidating the function of OsSYD and OsBRM during embryo development in rice.







Similar content being viewed by others
References
Bezhani S, Winter C, Hershman S, Wagner JD, Kennedy JF, Kwon CS, Pfluger J, Su Y, Wagner D (2007) Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell 19:403–416
Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78:273–304
Dellaporta S (1994) Plant DNA miniprep and microprep: versions 2.1–2.3. Springer, pp 522–525
Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86:423–433
Dresselhaus T (2006) Cell-cell communication during double fertilization. Curr Opin Plant Biol 9:41–47
Flaus A, Martin DM, Barton GJ, Owen-Hughes T (2006) Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res 34:2887–2905
Han SK, Wu MF, Cui S, Wagner D (2015) Roles and activities of chromatin remodeling ATPases in plants. Plant J 83:62–77
Hurtado L, Farrona S, Reyes JC (2006) The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol Biol 62:291–304
Ikeda-Kawakatsu K, Maekawa M, Izawa T, Itoh J, Nagato Y (2012) ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J 69:168–180
Ito M, Sentoku N, Nishimura A, Hong SK, Sato Y, Matsuoka M (2002) Position dependent expression of GL2-type homeobox gene, Roc1: significance for protoderm differentiation and radial pattern formation in early rice embryogenesis. Plant J 29:497–507
Ito M, Sentoku N, Nishimura A, Hong S-K, Sato Y, Matsuoka M (2003) Roles of rice GL2-type homeobox genes in epidermis differentiation. Breed Sci 53:245–253
Jin R, Klasfeld S, Zhu Y, Garcia MF, Xiao J, Han S-K, Konkol A, Wagner D (2021) LEAFY is a pioneer transcription factor and licenses cell reprogramming to floral fate. Nat Commun 12:1–14
Kamiya N, Nishimura A, Sentoku N, Takabe E, Nagato Y, Kitano H, Matsuoka M (2003) Rice globular embryo 4 (gle4) mutant is defective in radial pattern formation during embryogenesis. Plant Cell Physiol 44:875–883
Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T (2001) FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell 104:131–142
Khatun S, Flowers T (1995) The estimation of pollen viability in rice. J Exp Bot 46:151–154
King HW, Klose RJ (2017) The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. Elife 6:e22631
Kwon CS, Chen C, Wagner D (2005) WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev 19:992–1003
Kwon CS, Hibara K, Pfluger J, Bezhani S, Metha H, Aida M, Tasaka M, Wagner D (2006) A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development 133:3223–3230
Lai X, Blanc-Mathieu R, GrandVuillemin L, Huang Y, Stigliani A, Lucas J, Thevenon E, Loue-Manifel J, Turchi L, Daher H, Brun-Hernandez E, Vachon G, Latrasse D, Benhamed M, Dumas R, Zubieta C, Parcy F (2021) The LEAFY floral regulator displays pioneer transcription factor properties. Mol Plant 14:829–837
Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, Lahrmann U, Zhao Q, Zheng Y, Zhao Y, Xue Y, Ren J (2015) IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31:3359–3361
Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379:66–69
Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK (2013) Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant 6:2008–2011
Mayer U, Ruiz RAT, Berleth T, Miséra S, Jürgens G (1991) Mutations affecting body organization in the Arabidopsis embryo. Nature 353:402–407
Moyroud E, Tichtinsky G, Parcy F (2009) The LEAFY gloral regulators in angiosperms: conserved proteins with diverse roles. J Plant Biol 52:177–185
Mukherjee K, Brocchieri L, Burglin TR (2009) A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol Biol Evol 26:2775–2794
Nishimura A, Ito M, Kamiya N, Sato Y, Matsuoka M (2002) OsPNH1 regulates leaf development and maintenance of the shoot apical meristem in rice. Plant J 30:189–201
Palovaara J, de Zeeuw T, Weijers D (2016) Tissue and organ initiation in the plant embryo: a first time for everything. Annu Rev Cell Dev Biol 32:47–75
Pearson K (1900) X. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling. Lond Edinburgh Dublin Philos Magazine J Sci 50:157–175
Plackett AR, Conway SJ, Hewett Hazelton KD, Rabbinowitsch EH, Langdale JA, Di Stilio VS (2018) LEAFY maintains apical stem cell activity during shoot development in the fern Ceratopteris richardii. Elife 7:e39625
Sato Y, Hong S-K, Tagiri A, Kitano H, Yamamoto N, Nagato Y, Matsuoka M (1996) A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proc Natl Acad Sci 93:8117–8122
Sato Y, Sentoku N, Nagato Y, Matsuoka M (1998) Isolation and characterization of a rice homebox gene, OSH15. Plant Mol Biol 38:983–997
Sayou C, Nanao MH, Jamin M, Pose D, Thevenon E, Gregoire L, Tichtinsky G, Denay G, Ott F, Peirats Llobet M, Schmid M, Dumas R, Parcy F (2016) A SAM oligomerization domain shapes the genomic binding landscape of the LEAFY transcription factor. Nat Commun 7:11222
Sentoku N, Sato Y, Kurata N, Ito Y, Kitano H, Matsuoka M (1999) Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell 11:1651–1663
Shenghua W, Fang C, Kaida Z (2000) In vitro pollen germination of rice (Oryza sativa L.). Zuo Wu Xue Bao 26:609–612
Smith LG, Jackson D, Hake S (1995) Expression of knotted1 marks shoot meristem formation during maize embryogenesis. Dev Genet 16:344–348
Sugimoto N (1997) Temporal and spatial expression pattern of rice α-amylase gene Ramy1A during seed development. Rice Genet Newsl 14:148–150
Sun B, Zhou Y, Cai J, Shang E, Yamaguchi N, Xiao J, Looi L-S, Wee W-Y, Gao X, Wagner D, Ito T (2019) Integration of Transcriptional Repression and Polycomb-Mediated Silencing of WUSCHEL in Floral Meristems. Plant Cell 31:1488–1505
Tanahashi T, Sumikawa N, Kato M, Hasebe M (2005) Diversification of gene function: homologs of the floral regulator FLO/LFY control the first zygotic cell division in the moss Physcomitrella patens. Development 132:1727–1736
Tao Z, Shen L, Gu X, Wang Y, Yu H, He Y (2017) Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551:124–128
Truernit E, Haseloff J (2007) A role for KNAT class II genes in root development. Plant Signal Behav 2:10–12
Tsuda K, Ito Y, Sato Y, Kurata N (2011) Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23:4368–4381
Wagner D, Meyerowitz EM (2002) SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr Biol 12:85–94
Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69:843–859
Wu MF, Sang Y, Bezhani S, Yamaguchi N, Han SK, Li Z, Su Y, Slewinski TL, Wagner D (2012) SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc Natl Acad Sci 109:3576–3581
Xu L, Shen WH (2008) Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18:1966–1971
Xu P, Yuan D, Liu M, Li C, Liu Y, Zhang S, Yao N, Yang C (2013) AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots. Plant Physiol 161:1755–1768
Yang W, Gao M, Yin X, Liu J, Xu Y, Zeng L, Li Q, Zhang S, Wang J, Zhang X, He Z (2013) Control of rice embryo development, shoot apical meristem maintenance, and grain yield by a novel cytochrome P450. Mol Plant 6:1945–1960
Yi J, Lee Y-S, Lee D-Y, Cho M-H, Jeon J-S, An G (2016) OsMPK6 plays a critical role in cell differentiation during early embryogenesis in Oryza sativa. J Exp Bot 67:2425–2437
Acknowledgements
The authors thank Pro. Jiankang Zhu and Pro. Caixia Gao for offering the CRISPR/Cas9 gene-editing vector, and thank Pro. Junko Kyozuka for the gift of rfl/apo2 mutant seeds. We also appreciate the Biogle and Biorun Geneme Editing Center for producing transgenic rice.
Funding
This study was supported by the National Natural Science Foundation of China (Grant number 31872855 and 31971842).
Author information
Authors and Affiliations
Contributions
QX, CD and YD designed the research. QX, MM, YS, CR, JL, TZ and CD performed the experiments. QX and CD analyzed the data and prepared the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Accession numbers
Gene information in this article can be found in Rice Annotation Project database, miRbase data libraries and the Arabidopsis Information Resource are under the corresponding accession numbers: OsUBQ (Os03g0234200), OsSYD (Os06g0255200), RFL/APO2 (Os04g0598300), OsBRM (Os02g0114000), Roc1 (Os08g0187500), OsSCR (Os12g0122000), OsPNH1 (Os06g0597400), OSH1 (Os03g0727000), OSH6 (Os01g0302500), OSH15 (Os07g0129700), OSH71(Os05g0129700), RAmy1A(Os02g0765600).
Additional information
Communicated by Xian Sheng Zhang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xun, Q., Mei, M., Song, Y. et al. SWI2/SNF2 chromatin remodeling ATPases SPLAYED and BRAHMA control embryo development in rice. Plant Cell Rep 41, 1389–1401 (2022). https://doi.org/10.1007/s00299-022-02864-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-022-02864-z