Abstract
Key message
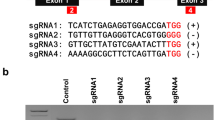
DCL2 and DCL4 genes in Nicotiana benthamiana plants were successfully edited using the CRISPR/Cas9 system.
Abstract
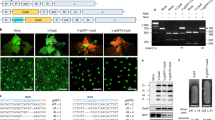
Recently, plants have been utilized for recombinant protein production similar to other expression systems, i.e., bacteria, yeast, insect, and mammal cells. However, insufficient amounts of recombinant proteins are often produced in plant cells. The repression of RNA silencing within plant cells could improve production levels of recombinant protein because RNA silencing frequently decomposes mRNAs from transgenes. In this study, the genes dicer-like protein 2 and 4 (NbDCL2 and NbDCL4) were successfully edited to produce double-knockout transgenic Nicotiana benthamiana plants (dcl2dcl4 plants) using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology. A transient green fluorescent protein (GFP) gene expression assay revealed that the dcl2dcl4 plants accumulated higher amounts of GFP and GFP mRNA than wild type (WT) and RNA-dependent RNA polymerase 6-knockout N. benthamiana plants (ΔRDR6 plants). Small RNA sequencing also showed that dcl2dcl4 plants accumulated lower amounts of small interfering RNAs (siRNAs) against the GFP gene than WT plants. The dcl2dcl4 plants might also produce higher amounts of human fibroblast growth factor 1 (FGF1) than WT and ΔRDR6 plants. These observations appear to reflect differences between DCLs and RDR6 in plant cell biological mechanisms. These results reveal that dcl2dcl4 plants would be suitable as platform plants for recombinant protein production.







Similar content being viewed by others
Availability of data
The raw sequence data reported are available in the DDBJ Sequence Read Archive (https://www.ddbj.nig.ac.jp/dra/index-e.html) under the accession number DRA009712.
References
Bhaskar PB, Venkateshwaran M, Wu L et al (2009) Agrobacterium-mediated transient gene expression and silencing: a rapid tool for functional gene assay in potato. PLoS ONE 4:e5812. https://doi.org/10.1371/journal.pone.0005812
Bouché N, Lauressergues D, Gasciolli V et al (2006) An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 25:3347–3356. https://doi.org/10.1038/sj.emboj.7601217
Chellappan P, Xia J, Zhou X et al (2010) siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res 38:6883–6894. https://doi.org/10.1093/nar/gkq590
Csorba T, Pantaleo V, Burgyán J (2009) RNA silencing: an antiviral mechanism. Adv Virus Res 75:35–71. https://doi.org/10.1016/S0065-3527(09)07502-2
Desai PN, Shrivastava N, Padh H (2010) Production of heterologous proteins in plants: strategies for optimal expression. Biotechnol Adv 28:427–435. https://doi.org/10.1016/j.biotechadv.2010.01.005
Drake PM, Szeto TH, Paul MJ et al (2017) Recombinant biologic products versus nutraceuticals from plants—a regulatory choice? Br J Clin Pharmacol 83:82–87. https://doi.org/10.1111/bcp.13041
Fahad S, Khan FA, Pandupuspitasari NS et al (2015) Recent developments in therapeutic protein expression technologies in plants. Biotechnol Lett 37:265–279. https://doi.org/10.1007/s10529-014-1699-7
Fernandez-Pozo N, Menda N, Edwards JD et al (2015) The Sol Genomics Network (SGN)–from genotype to phenotype to breeding. Nucleic Acids Res 43:D1036–D1041. https://doi.org/10.1093/nar/gku1195
Garabagi F, Gilbert E, Loos A et al (2012) Utility of the P19 suppressor of gene-silencing protein for production of therapeutic antibodies in Nicotiana expression hosts. Plant Biotechnol J 10:1118–1128. https://doi.org/10.1111/j.1467-7652.2012.00742.x
Gasciolli V, Mallory AC, Bartel DP et al (2005) Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15:1494–1500. https://doi.org/10.1016/j.cub.2005.07.024
Grosse-Holz F, Madeira L, Zahid MA et al (2018) Three unrelated protease inhibitors enhance accumulation of pharmaceutical recombinant proteins in Nicotiana benthamiana. Plant Biotechnol J 16:1797–1810. https://doi.org/10.1111/pbi.12916
Henderson IR, Zhang X, Lu C et al (2006) Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38:721–725. https://doi.org/10.1038/ng1804
Kościańska E, Kalantidis K, Wypijewski K et al (2005) Analysis of RNA silencing in agroinfiltrated leaves of Nicotiana benthamiana and Nicotiana tabacum. Plant Mol Biol 59:647–661. https://doi.org/10.1007/s11103-005-0668-x
Lico C, Santi L, Twyman RM et al (2012) The use of plants for the production of therapeutic human peptides. Plant Cell Rep 31:439–451. https://doi.org/10.1007/s00299-011-1215-7
Li JF, Norville JE, Aach J et al (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688–691. https://doi.org/10.1038/nbt.2654
Liu Y, Beyer A, Aebersold R (2016) On the dependency of cellular protein levels on mRNA abundance. Cell 165:535–550. https://doi.org/10.1016/j.cell.2016.03.014
Lombardi R, Circelli P, Villani ME et al (2009) High-level HIV-1 Nef transient expression in Nicotiana benthamiana using the P19 gene silencing suppressor protein of Artichoke Mottled Crinckle Virus. BMC Biotechnol 9:96. https://doi.org/10.1186/1472-6750-9-96
Lomonossoff GP, D’Aoust MA (2016) Plant-produced biopharmaceuticals: a case of technical developments driving clinical deployment. Science 353:1237–1240. https://doi.org/10.1126/science.aaf6638
Ludman M, Fátyol K (2019) The virological model plant, Nicotiana benthamiana expresses a single functional RDR6 homeolog. Virology 537:143–148. https://doi.org/10.1016/j.virol.2019.08.017
Ludman M, Burgyán J, Fátyol K (2017) Crispr/Cas9 mediated inactivation of argonaute 2 reveals its differential involvement in antiviral responses. Sci Rep 7:1010. https://doi.org/10.1038/s41598-017-01050-6
Lyon GM, Mehta AK, Unit VJB, Diseases ESC et al (2014) Clinical care of two patients with Ebola virus disease in the United States. N Engl J Med 371:2402–2409. https://doi.org/10.1056/NEJMoa1409838
Matsuo K (2021) Evaluation of methods for plant genomic DNA sequence analysis without DNA and PCR product purification. Plant Sci 312:111023. https://doi.org/10.1016/j.plantsci.2021.111023
Matsuo K, Atsumi G (2019) CRISPR/Cas9-mediated knockout of the RDR6 gene in Nicotiana benthamiana for efficient transient expression of recombinant proteins. Planta 250:463–473. https://doi.org/10.1007/s00425-019-03180-9
Matsuo K, Matsumura T (2017) Repression of the DCL2 and DCL4 genes in Nicotiana benthamiana plants for the transient expression of recombinant proteins. J Biosci Bioeng 124:215–220. https://doi.org/10.1016/j.jbiosc.2017.02.019
Matsuo K, Fukuzawa N, Matsumura T (2016) A simple agroinfiltration method for transient gene expression in plant leaf discs. J Biosci Bioeng 122:351–356. https://doi.org/10.1016/j.jbiosc.2016.02.001
Mitsuhara I, Ugaki M, Hirochika H et al (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37:49–59
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plantarum 15:473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Naito Y, Hino K, Bono H et al (2015) CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31:1120–1123. https://doi.org/10.1093/bioinformatics/btu743
Ocampo CG, Lareu JF, Marin Viegas VS et al (2016) Vacuolar targeting of recombinant antibodies in Nicotiana benthamiana. Plant Biotechnol J 14:2265–2275. https://doi.org/10.1111/pbi.12580
Parent JS, Bouteiller N, Elmayan T et al (2015) Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J 81:223–232. https://doi.org/10.1111/tpj.12720
Pillay P, Kibido T, du Plessis M et al (2012) Use of transgenic oryzacystatin-I-expressing plants enhances recombinant protein production. Appl Biochem Biotechnol 168:1608–1620. https://doi.org/10.1007/s12010-012-9882-6
Pombo MA, Ramos RN, Zheng Y et al (2019) Transcriptome-based identification and validation of reference genes for plant-bacteria interaction studies using Nicotiana benthamiana. Sci Rep 9:1632. https://doi.org/10.1038/s41598-018-38247-2
Rivard D, Anguenot R, Brunelle F et al (2006) An in-built proteinase inhibitor system for the protection of recombinant proteins recovered from transgenic plants. Plant Biotechnol J 4:359–368. https://doi.org/10.1111/j.1467-7652.2006.00187.x
Traynor K (2012) Taliglucerase alfa approved for Gaucher disease. Am J Health Syst Pharm 69:1009. https://doi.org/10.2146/news120041
Vazquez F, Blevins T, Ailhas J et al (2008) Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res 36:6429–6438. https://doi.org/10.1093/nar/gkn670
Webster GR, Teh AY, Ma JK (2017) Synthetic gene design–The rationale for codon optimization and implications for molecular pharming in plants. Biotechnol Bioeng 114:492–502. https://doi.org/10.1002/bit.26183
Yang J, Barr LA, Fahnestock SR et al (2005) High yield recombinant silk-like protein production in transgenic plants through protein targeting. Transgenic Res 14:313–324. https://doi.org/10.1007/s11248-005-0272-5
Ye R, Chen Z, Lian B et al (2016) A Dicer-independent route for biogenesis of siRNAs that direct dna methylation in Arabidopsis. Mol Cell 61:222–235. https://doi.org/10.1016/j.molcel.2015.11.015
Zhang K, Raboanatahiry N, Zhu B et al (2017) Progress in genome editing technology and its application in plants. Front Plant Sci 8:177. https://doi.org/10.3389/fpls.2017.00177
Acknowledgements
I thank Dr. Go Atsumi and Dr. Takeshi Matsumura for their advice. This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant no. 16K14833).
Funding
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant no. 16K14833).
Author information
Authors and Affiliations
Contributions
KM conceived and designed the work. KM secured funding. KM performed the experiments. KM analyzed the data. KM drafted and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The author has no conflicts of interest to declare.
Additional information
Communicated by Fumihiko Sato.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
299_2021_2809_MOESM1_ESM.tif
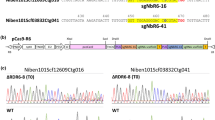
Supplementary Fig. S1 Mutations in the NbDCL2 and NbDCL4 alleles of transgenic line D24-10 (T0). DNA fragments around the target sequences were amplified by PCR and then subjected to sequencing. PAM (protospacer adjacent motif) and the sgRNA target sequences are indicated by red and blue lines, respectively. Red arrows indicate inserted nucleotides (TIF 1426 KB)
299_2021_2809_MOESM2_ESM.tif
Supplementary Fig. S2 Mutations in the NbDCL2 and NbDCL4 alleles of transgenic line D24-10-AH (T1). DNA fragments around the target sequences were amplified by PCR and then subjected to sequencing. PAM (protospacer adjacent motif) and the sgRNA target sequences are indicated by red and blue lines, respectively. Red arrows indicate inserted nucleotides (TIF 1521 KB)
299_2021_2809_MOESM3_ESM.tif
Supplementary Fig. S3 Mutations in the NbDCL2 and NbDCL4 alleles of transgenic line D24-10-AH-3 (T2). DNA fragments around the target sequences were amplified by PCR and then subjected to sequencing. PAM (protospacer adjacent motif) and the sgRNA target sequences are indicated by red and blue lines, respectively. Red arrows indicate inserted nucleotides (TIF 1452 KB)
299_2021_2809_MOESM4_ESM.tif
Supplementary Fig. S4 Off-target analysis of dcl2dcl4 plants. The PAM and the potential off-target site matching the 12 bp sequence of the target sequence is indicated by red and blue lines, respectively (TIF 647 KB)
Rights and permissions
About this article
Cite this article
Matsuo, K. CRISPR/Cas9-mediated knockout of the DCL2 and DCL4 genes in Nicotiana benthamiana and its productivity of recombinant proteins. Plant Cell Rep 41, 307–317 (2022). https://doi.org/10.1007/s00299-021-02809-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-021-02809-y