Abstract
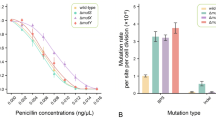
Mutations in diploid budding yeast occur in meiosis at higher frequencies than in cells grown vegetatively. Such meiotic mutations are thought to result from the repair of double-strand breaks (DSBs) in meiosis, during the process of recombination. Here, we report studies of mutagenicity in haploid strains that may undergo meiosis due to the expression of both mating-type alleles, MATa and MATα. We measure the rate of mutagenicity in the reporter gene CAN1, and find it to be fivefold higher than in mitotic cells, as determined by fluctuation analysis. This enhanced meiotic mutagenicity is shown to depend on the presence of SPO11, the gene responsible for meiotic DSBs. Mutations in haploid meiosis must result from repair of the DSBs through interaction with the sister chromatid, rather than with non-sister chromatids as in diploids. Thus, mutations in diploid meiosis that are not ostensibly associated with recombination events can be explained by sister-chromatid repair. The spectrum of meiotic mutations revealed by Sanger sequencing is similar in haploid and in diploid meiosis. Compared to mitotic mutations in CAN1, long Indels are more frequent among meiotic mutations. Both, meiotic and mitotic mutations are more common at G/C sites than at A/T, in spite of an opposite bias in the target reporter gene. We conclude that sister-chromatid repair of DSBs is a major source of mutagenicity in meiosis.
Similar content being viewed by others
Availability of data and material
All reagents are available upon request.
References
Alani E, Cao L, Kleckner N (1987) A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541–545
Arbel A, Zenvirth D, Simchen G (1999) Sister chromatid-based repair is mediated by RAD54, not by DMC1 or TID1. EMBO J 18:2648–2658
Arbel-Eden A, Simchen G (2019) Elevated mutagenicity in meiosis and its mechanism. BioEssays 41:272–281
Bergerat A, De Massy B, Baudat F et al (1997) An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386:414–417
De Massy B, Baudat F, Nicolas A (1994) Initiation of recombination in Saccharomyces cerevisiae haploid meiosis. Acad Sci USA 91:11929–11933
Dov A (2015) The contribution of trans-lesion DNA polymerases Rev1, Rev3, Rad30 to the meiotic mutagenesis of budding yeast. MSc Dissertation, The Hebrew University of Jerusalem, Jerusalem (Accession No. H/JSL/002000812). In Hebrew
Goldfarb T, Lichten M (2010) Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol 8(10):e1000520
Haag-Liautard C, Dorris M, Maside X et al (2007) Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445:82–85
Hugerat Y, Simchen G (1993) Mixed segregation and recombination of chromosomes and YACs during single-division meiosis in spo13 strains of Saccharomyces cerevisiae. Genetics 135:297–308
Kassir Y, Simchen G (1991) Monitoring meiosis and sporulation in Saccharomyces cerevisiae. Methods Enzym 194:94–110
Kassir Y, Granot D, Simchen G (1988) IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell 52:853–862
Katis VL, Matos J, Mori S et al (2004) Spo13 facilitates monopolin recruitment to kinetochores and regulates maintenance of centromeric cohesion during yeast meiosis. Curr Biol 14:2183–2196
Keeney S (2001) Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol 52:1–53
Keeney S, Neale MJ (2006) Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem Soc Trans 34:523–525
Keeney S, Giroux CN, Kleckner N (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88:375–384
Kerr GW, Sarkar S, Arumugam P (2012) How to halve ploidy: lessons from budding yeast meiosis. Cell Mol Life Sci 69:3037–3051
Klapholz S, Esposito RE (1980) Isolation of SPO12-1 and SPO13-1 from a natural variant of yeast that undergoes a single meiotic division. Genetics 96:567–588
Lang GI, Murray AW (2008) Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 178:67–82
Lee BH, Amon A, Prinz S (2002) Spo13 regulates cohesin cleavage. Genes Dev 16:1672–1681
Loidl J, Nairz K (1997) Karyotype variability in yeast caused by nonallelic recombination in haploid meiosis. Genetics 146:70–88
Luria SE, Delbrück M (1943) Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491–511
Magni GE (1963) The origin of spontaneous mutations during meiosis. Proc Natl Acad Sci USA 50:975–980
Magni GE (1964) Origin and nature of spontaneous mutations in meiotic organisms. J Cell Physiol 64:65–71
Magni GE, Von Borstel RC (1962) Different rates of spontaneous mutation during mitosis and meiosis in yeast. Genetics 47:1097–1108
Mansour O, Morciano L, Zion K et al (2020) Timing of appearance of new mutations during yeast meiosis and their association with recombination. Curr Genet 66:577–592
Ness RW, Morgan AD, Colegrave N et al (2012) Estimate of the spontaneous mutation rate in Chlamydomonas reinhardtii. Genetics 192:1447–1454
Prinz S, Amon A, Klein F (1997) Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 146:781–795
Rattray A, Santoyo G, Shafer B et al (2015) Elevated mutation rate during meiosis in Saccharomyces cerevisiae. PLoS Genet 11:e100491
Rosche WA, Foster PL (2000) Determining mutation rates in bacterial populations. Methods 20:4–17
Rose M, Winston F, Hieter P (1990) Methods in yeast genetics - a laboratory course manual. Cold Spring Harbor, New York
Sharon G, Simchen G (1990) Mixed segregation of chromosomes during single-division meiosis of Saccharomyces cerevisiae. Genetics 125:475–485
Shonn MA, McCarroll R, Murray AW (2002) Spo13 protects meiotic cohesin at centromeres in meiosis I. Genes Dev 16:1659–1671
Simchen G, Hugerat Y (1993) What determines whether chromosomes segregate reductionally or equationally in meiosis? BioEssays 15:1–8
Wagstaff JE, Klapholz S, Esposito RE (1982) Meiosis in haploid yeast. Proc Natl Acad Sci USA 79:2986–2990
Whelan WL, Gocke E, Manney TR (1979) The CAN1 locus of Saccharomyces cerevisiae: fine-structure analysis and forward mutation rates. Genetics 91:35–51
Humphryes N, Hochwagen A (2014) A non-sister act: recombination template choice during meiosis. Exp Cell Res 329:53–60
Acknowledgements
We thank our colleague Dr. Tanvi Suhane for her useful comments on the manuscript.
Funding
Supported by funds provided by the Israel Science Foundation (Grants No. 501/16 and 397/20), the US-Israel Binational Science Foundation (Grant No. 2009299) and by Hadassah Academic College.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: GS, AAE. Performed the experiments: OM, LM. Contributed reagents/materials/analysis tools: DZ, LM. Wrote the paper: GS.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest or competing interests regarding this work.
Additional information
Communicated by Michael Polymenis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Simchen, G., Mansour, O., Morciano, L. et al. Mutagenicity in haploid yeast meiosis resulting from repair of DSBs by the sister chromatid. Curr Genet 67, 799–806 (2021). https://doi.org/10.1007/s00294-021-01189-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-021-01189-w