Abstract
Since its description in S. cerevisiae, the Regulation of Ace2 and Morphogenesis (RAM) pathway has been studied for nearly 20 years in multiple model and pathogenic fungi. In pathogenic fungi, the RAM pathway carries out many functions through mechanisms that remain to be defined in detail. Recently, we reported that Cbk1-mediated phosphorylation of the transcription factor Ace2 functions to repress the hyphae-to-yeast transition in Candida albicans. This transition is understudied relative to the yeast-to-hyphae transition. Subapical hyphal cell compartments are arrested in G1 until the point at which lateral yeast emerge. Here, we discuss this model and report new data indicating that a second G1 associated protein, the mitotic exit regulator Amn1. In S. cerevisiae diploid cells, Amn1 negatively regulates Ace2 at both the gene expression level through a negative feedback loop and at the protein level by targeting Ace2 for degradation. In C. albicans, Amn1 and Ace2 also form a feedback loop at the level of gene expression. Deletion of AMN1 decreases lateral yeast formation relative to wild type in maturing hyphae and is associated with decreased expression of PES1, a positive regulator of lateral yeast formation. These data indicate that the regulation of mitotic exit plays a role in determining the timing of lateral yeast emergence from hyphae in C. albicans. We also propose an integrated model for the interplay between the Cbk1-Ace2 axis and other hyphal stage regulators during the process of filamentation and transition back to yeast.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Candida albicans is an important human fungal pathogen that causes both superficial, mucosal infections and deep organ, invasive disease in both immunocompetent and immunocompromised individuals (McCarty and Pappas 2016). C. albicans is also part of the normal human mycobiome with primary niches in the gastrointestinal and genitourinary tracts (Romo and Kumamoto 2020). During infection, C. albicans adopts at least three distinct morphological cell types: the familiar budding yeast form; pseudohyphae; and true hyphae (Fig. 1a, for detailed definitions of each morphotype see: Sudbery 2011). Because the ability to transition between these morphotypes affects the virulence of C. albicans, the processes that regulate these transitions have been the subject of intensive study for decades (Arkowitz and Bassilana 2020). The vast majority of these studies have focused on the transition from yeast to hyphae or pseudohyphae because this transition is most tightly correlated with virulence in mammals.
A significantly understudied transition occurs after a hyphal cell has accumulated multiple sub-apical cell compartments. At this stage, yeast cells re-emerge from the hyphae and thus a hyphae-to-yeast transition occurs (Shen et al. 2008). These sub-apical cells are typically referred to as lateral yeast cells (Fig. 1b). Increased interest in the hyphae to yeast transition has been driven in part by the relatively recent recognition that it plays an important role in biofilm pathobiology (Uppuluri et al. 2010). Specifically, the outer layers of C. albicans biofilms release yeast cells or dispersed cells as the structure matures. Dispersed cells are thought to mediate dissemination of C. albicans from biofilms to other parts of the body and, thus, play an important role in the infection process (Wall et al. 2019).
Lateral yeast formation is delayed until several rounds of hyphal cell extension have occurred (Fig. 1c). The mother cells and sub-apical compartments are arrested in G1 while the apical compartment is mitotically active (Sudbery 2011). Gow and co-workers developed a model to explain the G1 arrest that is based on the observation that newly formed hyphal compartments are highly vacuolated and thus have relatively reduced cytoplasmic volume (Gow and Gooday 1987). This low cytoplasm content means that the hyphal compartments are equivalent to much smaller cells—i.e., new, small daughter cells. Cell size is a well-characterized determinant of cell cycle re-entry in yeast (Chaillot et al. 2017). Once the daughter cell has grown to a specific size it completes its transition from G1 to START. As the sub-apical compartments mature, the cytoplasm/vacuole ratio increases to the point where the cells exit from G1 and enter START (Barelle et al. 2003), leading to the emergence of lateral yeast cells from subapical compartments. Although this cell biological model nicely fits the observed data, almost nothing is known about the molecular details underlying the mechanism of the hyphae-to-yeast transition.
An important exception to this general statement comes from Kohler and co-workers who found that expression of the pescadillo homologue PES1 is both necessary and sufficient to drive lateral yeast formation (Shen et al. 2008). In C. albicans, PES1 is essential in yeast cells but dispensable in hyphae. Overexpression of PES1 increases the formation of lateral yeast while decreased expression using a doxycycline-regulated allele reduced lateral yeast formation. In addition, PES1 has been shown to play a role in the formation of biofilm dispersal cells.
How Pes1 regulates the hyphae to yeast transition remains unknown. In a second study, Lindsay et al. reported that farnesol induces lateral yeast formation (Lindsay et al. 2012). Farnesol inhibits hyphal morphogenesis by decreasing the activity of the Ras1-cAMP pathway. These data suggest that the emergence of lateral yeast is dependent on a down-regulation of the Ras1-cAMP pathway (Lindsay et al. 2012). Although these studies provide important initial clues to the regulation of the hyphae to yeast transition, much more remains to be learned. Here, we discuss our recent work on the role of the Regulation of Ace2 and Morphogenesis (RAM) pathway in the regulation of hyphal-related processes (Wakade et al. 2020) and present data providing new details regarding the role of the mitotic exit regulator Amn1 in the modulation of ACE2 expression and the formation of lateral yeast cells from hyphae.
Results and discussion
The RAM pathway (Fig. 2a) was initially described in S. cerevisiae and is conserved across both pathogenic and model fungi (Nelson et al. 2003; Saputo et al. 2012). The RAM pathway is made up of a Ndr/Lats kinase (Cbk1 in C. albicans and S. cerevisiae) along with an additional kinase Kic1 as well as four binding partners (Hym1, Sog2, Tao3/Pag1, and Mob2). The latter five proteins are required to activate Cbk1 with Mob2 directly binding Cbk1. The transcription factor Ace2 was incorporated into the pathway’s name because it was the first effector molecule shown to be dependent on the RAM pathway (Nelson et al. 2003). The most extensively studied function of the Cbk1-Ace2 axis is its role in daughter cell specific expression of septum-degrading genes required for cell separation (Sbia et al. 2008). Cbk1 phosphorylates Ace2 near its nuclear export signal during G1, trapping it in the nucleus (Mazanka et al. 2008). The Weiss lab established that Ace2 mutants lacking Cbk1 phospho-acceptor residues in S. cerevisiae display cell separation defects and have reduced expression of cell septum degrading enzymes such as chitinases and glucanases. Recently, we generated the analogous Cbk1 phosphosite mutants of Ace2 in C. albicans and showed that, as in S. cerevisiae, Cbk1 phosphorylation of Ace2 is required for the accumulation of Ace2 in the nuclei of daughter cells in yeast phase growth, expression of chitinases and glucanases, and ultimately, normal mother-daughter cell separation (Wakade et al. 2020).
Regulation of Ace2 and Morphogenesis (RAM) pathway and Cbk1 phosphosites of Ace2. a Schematic showing the different components of the RAM pathway and examples of three Cbk1-regulated transcription factors in C. albicans. b The location of Cbk1 consensus phosphorylation sites and the Cbk1 dependent functions that were identified by mutation of those sites to non-phosphorylatable alanine (Wakade et al. 2020)
C. albicans Cbk1 and, under specific conditions, Ace2 are also involved in filamentous growth. Cbk1 is absolutely required for filamentous growth due, at least in part, to its role in the negative regulation of Nrg1, a key repressor of the hyphal program (Lee et al. 2015). The best characterized role for Ace2 during filamentation is under hypoxic and embedded conditions where it is required for normal hyphal morphogenesis (Mulhern et al. 2006; Desai et al. 2016). We found that Cbk1 phosphorylation of Ace2 plays a modest role in filamentation under hypoxic conditions but no role when cells are embedded within agar (Wakade et al. 2020). Deletion of ACE2 has a minimal effect on in vitro filamentation using standard liquid induction conditions such as serum (Saputo et al. 2014), Spider medium or other medium and we confirmed that was the case for strains lacking Cbk1 phosphorylation sites (Wakade et al. 2020).
During these experiments (Wakade et al. 2020), we noticed that ace2∆∆ strains developed lateral yeast cells at time points when the WT comparator had very few such cells. Specifically, after 4 h of induction in Spider medium at 37 °C, only 10% of hyphae formed by the SN250 reference strain displayed lateral yeast cells while 40% of hyphae formed by the congenic ace2∆∆ strain had formed lateral yeast cells. Mutation of the Cbk1 phosphorylation sites to alanine increases lateral yeast formation to levels (50%) that closely match the ace2∆∆ null mutant, indicating that Cbk1 phosphorylation of Ace2 leads to suppression of lateral yeast cell formation during hyphal morphogenesis (Wakade et al. 2020).
These results have novel implications for the function of Cbk1-Ace2 and for the regulation of lateral yeast formation. First, this is only the second regulatory pathway linked to the timing of lateral yeast formation (Lindsay et al. 2012). Second, Ace2 must have functions outside of the new daughter/apical cell during hyphal morphogenesis and thus functions in the subapical compartments of the hyphae. We and others have shown that Ace2 localizes to the apical cell nuclei of C. albicans hyphae but it is not detectable in either the cytoplasm or nuclei of sub-apical cells (Kelly et al. 2004; Bharucha et al. 2011). Paradoxically, despite being predominantly localized to the apical cell nuclei, Ace2 has no effect on the emergence of the new hyphal cell body while it has a significant effect on the biology of sub-apical cells in which it appears to be present at vanishingly low levels.
As mentioned above, PES1 is necessary, and sufficient, for lateral yeast formation (Shen et al. 2008) and, consistent with this role, its expression is upregulated in ace2∆∆ deletion mutants relative to wild type hyphal cells (Wakade et al. 2020). At this point, it is not clear whether Ace2 directly suppresses PES1 transcription or not. Our RNA sequencing data shows that a large set of genes (144) is upregulated in hyphal ace2∆∆ cells compared to wild type (Wakade et al. 2020); however, there are no biochemical data indicating that Ace2 is a direct repressor of gene transcription nor have chromatin immunoprecipitation experiments indicated that Ace2 binds to the promoter of PES1 (Desai et al. 2016). Still, many transcription factors function as both activators and repressors.
Regardless of the specific mechanism by which loss Ace2 function triggers lateral yeast formation, the early development of lateral yeast does not affect the ongoing hyphae formation. The hyphae formed by ace2∆∆ are otherwise unchanged (Mulhern et al. 2006; Saputo et al. 2014). Furthermore, hypha-specific genes such as HWP1 are expressed at normal or slightly increased levels in ace2∆∆ cells (Wakade et al. 2020). Consequently, both hyphal and yeast phase growth programs are operative over the length of the hyphae. As such, although Cbk1-activated Ace2 prevents the premature resumption of yeast cell morphogenesis, its role is distinct from factors that function in hyphal maintenance (Lu et al. 2014) because the hyphae program continues in its absence.
The function of Ace2 in the subapical cells is consistent with its cell cycle role as a G1 factor in S. cerevisiae (Sbia et al. 2008; Mazaka and Weiss 2010). Indeed, Ace2 has been proposed to delay G1 in daughter cells based on overexpression studies (Laabs et al. 2003). However, others have shown that the role of Ace2 may be more complex because deletion of ACE2 appears to equalize the growth rate of mother and daughter cells rather than accelerate the cell cycle (Bogomolnaya et al. 2006). As mentioned earlier, the subapical cells of C. albicans are thought to be arrested in G1 (Gow and Gooday 1987; Sudbery 2011) and hence it seems likely that active Ace2 would be present in those cell compartments. Recently, the S. cerevisiae mitotic exit regulator Amn1 was reported to bind Ace2 and target it for ubiquitin-dependent degradation (Fang et al. 2018). Interestingly, Ace2 is also required for expression of AMN1, but only in S. cerevisiae diploid cells and not haploids. We were, therefore, interested to see if Amn1 played a role in the regulation of ACE2 expression or lateral yeast formation.
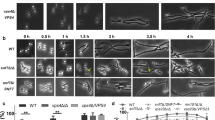
To explore the relationship between Ace2 and Amn1 further, we examined the expression of AMN1 in ace2∆∆ during hyphae formation in our previously reported RNA seq data (Wakade et al. 2020). Indeed, AMN1 expression is reduced in ace2∆∆ yeast cells hyphae relative to WT hyphae and we confirmed this in hyphal cells by RT-PCR (Fig. 3a). If Amn1 negatively regulates Ace2 function in C. albicans, then deletion of AMN1 should lead to increased expression of Ace2 targets such as CHT3 and SCW11 and, in fact, this is the case (Fig. 3b). Consistent with the increased Ace2 function in amn1∆∆ mutants, these strains show decreased lateral yeast formation relative to wild type cells in Spider medium at both 4 and 8 h post-hyphal induction (Fig. 3c). The hyphae of amn1∆∆ strains are otherwise normal true hyphae (Fig. 3d). We previously showed that PES1 expression is increased in ace2∆∆ strains. In keeping with the reduced lateral yeast formation in amn1∆∆ cells, PES1 expression is reduced in the hyphae of amn1∆∆ mutants relative to wild type (Fig. 3e).
Transcriptional interplay between Ace2 and the G1 regulator Amn1 in C. albicans. a AMN1 expression is decreased in ace2∆∆ in both yeast and hyphal phase as determined by RNA-sequencing experiments (Wakade et al. 2020). The effect was confirmed for hyphae using RT-PCR. b Deletion of AMN1 increases the expression of Ace2 targets CHT3 and SCW11 in hyphae. c Lateral yeast emergence is delayed in amn1∆∆ mutants relative to wild type following hyphal induction in Spider medium at 37 °C. d Micrographs of WT and amn1∆∆ strains induced in Spider medium as for (c) showing that the deletion mutant forms true hyphae. e The expression of PES1 was determined by RT-PCR 4 and 8 h after hyphal induction with Spider medium at 37 °C. For a, b and e bars indicate the mean ratio of the expression of the indicated gene expressed as fold-change in the ratio of mutant/wild type. Error bars indicate the standard deviation of two biological replicates performed in duplicate. Significant differences were determined by Student’s t test with a threshold of P < 0.05: *P < 0.05; **P < 0.01; ***P < 0.001. Bars indicate the number of hyphae with lateral yeast as determined by light microscopy. For c, at least 100 cells were characterized per replicate. The error bars indicate the standard deviation for at least two independent experiments. Student’s t test analysis of the difference between the wild type and amn1∆∆ cells indicated it was statistically significant (P < 0.05)
In S. cerevisiae, Amn1 binds Ace2 and targets it to the E3 ligase which in turn leads to proteasomal degradation but has essentially no effect on the expression of ACE2 RNA (Fang et al. 2018). To investigate the mechanism by which Amn1 affects Ace2 function during hyphae formation, we first compared the expression of ACE2 in wt and amn1∆∆ strains after hyphal induction by qRT-PCR. Surprisingly, we found that ACE2 expression is increased in amn1∆∆ by three-fold (Fig. 4a), indicating that Amn1 plays a role in the regulation of ACE2 expression. Amn1, however, has no DNA binding sequences and, thus, its regulation of ACE2 expression must be indirect. However, this increased expression would explain the decreased PES1 expression in amn1∆∆ strains. As noted above, the effect of Amn1 on ACE2 expression is distinct from the case in diploid S. cerevisiae cells under yeast phase growth. Additional experiments will be needed to dissect the relative contributions of gene expression and possible protein degradation in the role of Amn1 in ACE2 steady state levels during hyphae formation.
Amn1 and Ace2 form a feedback loop at the transcriptional level and regulate PES1 expression. a ACE2 expression is increased in amn1∆∆ mutants during hyphae induction. Bars indicate the mean ratio of the expression of the indicated gene expressed as fold-change in the ratio of mutant/wild type. Error bars indicate the standard deviation of two biological replicates performed in duplicate. Significant differences were determined by Student’s t test with a threshold of P < 0.05: **P < 0.01. b Data from Fig. 3a/4a and previously published data showing that Ace2 represses PES1 expression during hyphae formation (Wakade et al. 2020) provides genetic support for the indicated feedback loop
Importantly, our data indicate that, unlike S. cerevisiae (Fang et al. 2018), there is a direct or indirect mechanism by which Amn1 inhibits the transcription of ACE2 in C. albicans. It is possible that Amn1 also binds and degrades other Ace2-regulating transcription factors, such as Tec1 or Brg1. Since Ace2 induces the expression of Amn1 and Amn1 reduces the expression of Ace2, there appears to be a negative feedback loop that links Amn1 levels to ACE2 expression (Fig. 4b). Genome-wide chromatin immunoprecipitation data have not found that Ace2 binds its own promoter (Desai et al. 2016); although this does not rule out autoregulation of ACE2, it suggests that other mechanisms may be operative. Ace2 does, however, bind to the promoter of BRG1 (Desai et al. 2016) and, as we have shown, Brg1 binds to the ACE2 promoter and is required for full expression ACE2 expression during hyphae formation (Saputo et al. 2014; Desai et al. 2016). Thus, we suggest that Ace2 and Brg1 form a positive feedback loop that regulates the increase in ACE2 expression after initiation of hyphal development in C. albicans. The steady state levels of ACE2 expression, therefore, are determined by the relative activities of these two competing, feedback loops.
Based on these new data and previously reported results, we propose an integrated model for the regulation of ACE2 expression (Fig. 5). During hyphal induction, the levels of the hyphal repressor Nrg1 are reduced in a Cbk1-dependent process (Lee et al. 2015). Nrg1 represses the expression of Brg1 and relief of that repression occurs early in hyphal induction (Cleary et al. 2012) leading to a dramatic increase in Brg1 expression which, along with Tec1, increases ACE2 expression by initiating a positive feedback loop (Saputo et al. 2014; Finkel et al. 2012). This positive feedback loop appears to be balanced by a negative feedback loop through the Ace2 driven expression of AMN1. The Ace2-Amn1 negative feedback loop operates indirectly at the transcriptional level but may occur at the protein level as well.
Early in hyphal induction the two competing feedback loops are shifted toward a net increase in ACE2 expression (Saputo et al. 2014). The sub-apical hyphal compartments are arrested in G1 and Ace2 expression remains high enough to suppress lateral yeast formation. After approximately 3–4 h of hyphal induction, Nrg1 expression increases and, consequently, BRG1 expression is reduced (Cleary et al. 2012). Based on these data, we propose that the positive feedback loop maintaining ACE2 expression is interrupted and the balance tips toward Amn1-mediated reduction in Ace2 levels. Once the negative feedback loop reduces Ace2 levels to a critical level, G1-START is initiated and lateral yeast cells then begin to emerge from the hyphae.
One of the challenges in studying these events is that hyphal development cannot be synchronized. Thus, it is important to keep in mind that the transcriptional and protein levels measured for these processes are bulk averages for cultures containing cells at varying stages of development. Time course experiments for ACE2 gene expression show that expression peaks at 3 h and is decreasing by 5 h with changes in protein levels lagging behind gene expression (Saputo et al. 2014). Thus, within the limits of experiments using bulk cultures, the timing of increased NRG1 expression and decreasing ACE2 expression fits reasonably well with the beginning of lateral yeast formation. The development of single cell/hyphae reporters of the proposed events would allow high resolution testing of this model.
Additional work is required to place this model on a firm biochemical foundation but its general features are consistent with the current genetic and transcriptional data and generate hypotheses for further testing. Specifically, a number of mechanistic questions remain unanswered, including: Is Ace2 acting as a transcriptional repressor in the G1 arrested hyphal compartments? If so, does it directly regulate PES1? If not, then how is it repressing the emergence of lateral yeast? And finally, do similar processes occur during the emergence of dispersed yeast cells from biofilms? With respect to the latter question, AMN1 expression is increased in mature in vivo biofilms relative to initial biofilms (Nett et al. 2009), suggesting that AMN1 may have a role in dispersal of yeast cells from biofilms.
Our findings that two G1-related proteins, Ace2 and Amn1, affect the timing of the hyphae-to-yeast transition are consistent with previous cell biological model based on delayed G1. These data provide some of the first information regarding the molecules that are involved in regulating the G1 arrest in subapical cells of C. albicans hyphae. In summary, our work and that of others has clearly shown that the Cbk1-Ace2 axis plays a unique role in C. albicans biology in that it regulates aspects of both the initiation and termination of hyphal morphogenesis.
Methods
Strains, media, and cultivation
All strains are derivatives of the SN background (Noble and Johnson 2005) and were constructed by deletion of ACE2 or AMN1 using the transient CRISPR/Cas9 method (Min et al. 2016). Homozygous integration of the deletion cassette was confirmed by standard PCR methods as previously described (Wakade et al. 2020). Yeast peptone dextrose (YPD) and Spider medium (SM) were prepared using standard recipes (Homann et al. 2009). Strains were pre-cultured in YPD overnight at 30 °C. For yeast phase growth, the cultures were back-diluted to a cell density of 0.05–0.1 OD600 and incubated at 30 °C until the density had reached between 0.5 and 1 OD600. For hyphal induction, the overnight culture was diluted 1:50 in SM and shifted to 37 °C for induction.
Quantitative reverse transcription-PCR (qPCR)
RNA was isolated from yeast and hyphal phase cells using RiboPure ™ kit and reverse transcribed using an iScript cDNA synthesis kit (170-8891; Bio-Rad). The qPCR reaction was performed using IQ SyberGreen supermix (170-8882; Bio-Rad). Briefly, each reaction contained 10 μ of the SYBER Green PCR master mix, 0.10 μM of the respective primers and 150 ng of cDNA as a template in a total volume of 20 μL. Data analysis was performed using 2−ΔΔCT method and ACT1 was used as an internal control. Data reported here are the means from 3 independent biological replicates performed in a triplicate.
Characterization of lateral yeast formation
SN250 or amn1∆∆ strains were induced to form hyphae as described above. The cells were harvested after incubation for 4 and 8 h, fixed, and then examined by light microscopy. The number of hyphae with lateral yeast cells was determined for each strain by counting at least 100 hyphae for each replicate. The data are reported as the percentage of hyphae with lateral yeast cells and are means of two independent experiments. Differences between the reference SN250 and amn1∆∆ were analyzed by Student’s t test and the P value was less than 0.05 indicating statistical significance.
References
Arkowitz RA, Bassilana M (2020) Recent advances in understanding Candida albicans hyphal growth. F1000Res 21:8
Barelle CJ, Bohula EA, Kron SJ, Wessels D, Soll DR, Schäfer A, Brown AJ, Gow NA (2003) Asynchronous cell cycle and asymmetric vacuolar inheritance in true hyphae of Candida albicans. Eukaryot Cell 2:398–410
Bharucha N, Chabrier-Rosello Y, Xu T, Johnson C, Sobczynski S, Song Q, Dobry CJ, Eckwahl MJ, Anderson CP, Benjamin AJ, Kumar A, Krysan DJ (2011) A large-scale complex haploinsufficiency-based genetic interaction screen in Candida albicans: analysis of the RAM network during morphogenesis. PLoS Genet 7:e1002058
Bogomolnaya LM, Pathak R, Guo J, Polymenis M (2006) Roles of the RAM signaling network in cell cycle progression in Saccharomyces cerevisiae. Curr Genet 49:384–392
Chaillot J, Cook MA, Corbeil J, Sellam A (2017) Genome-wide screen for haploinsufficient cell size genes in the opportunistic yeast Candida albicans. G3 (Bethesda) 7:355–360
Cleary IA, Lazzell AL, Monteagudo C, Thomas DP, Saville SP (2012) Brg1 and Nrg1 form a novel feedback circuit regulating Candida albicans hypha formation and virulence. Mol Microbiol 85:557–573
Desai PR, van Wijlick L, Kurtz D, Juchimiuk M, Ernst JF (2016) Hypoxic and temperature regulated morphogenesis in Candida albicans. PLoS Genet 11:e1005447
Fang O, Hu X, Wang L, Jiang N, Yang J, Li B, Luo Z (2018) Amn1 governs post-mitotic separation in Saccharomyces cerevisiae. PLoS Genet 14:e1007691
Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, Nett JE, Taff H, Norice CT, Andes DR, Lanni F, Mitchell AP (2012) Portrait of Candida albicans adherence regulators. PLoS Pathog 8:e1002525
Gow NA, Gooday GW (1987) Cytological aspects of dimorphism in Candida albicans. Crit Rev Microbiol 15:73–78
Homann OR, Dea J, Noble SM, Johnson AD (2009) A phenotypic profile of the Candida albicans regulatory network. PLoS Genet 5:e1000783
Kelly MT, MacCallum DM, Clancy SD, Odds FC, Brown AJ, Butler G (2004) The Candida albicans CaACE2 gene affects morphogenesis, adherence, and virulence. Mol Microbiol 53:969–983
Laabs TL, Markwardt DD, Slattery MG, Newcomb LL, Stillman DJ, Heideman W (2003) ACE2 is required for daughter cell-specific G1 delay in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 100:10275–10280
Lee HJ, Kim JM, Kang WK, Yang H, Kim JY (2015) The NDR kinase Cbk1 downregulates the transcriptional repressor Nrg1 through the mRNA-binding protein Ssd1 in Candida albicans. Eukaryot Cell 14:671–683
Lindsay AK, Deveau A, Piispanen AE, Hogan DA (2012) Farnesol and cyclic AMP signaling effects on the hypha-to-yeast transition in Candida albicans. Eukaryot Cell 11:1219–1225
Lu Y, Su C, Liu H (2014) Candida albicans hyphal initiation and elongation. Trends Microbiol 22:707–714
Manzaka E, Weiss EL (2010) Sequential counteracting kinases restrict an asymmetric gene expression program to early G1. Mol Biol Cell 19:2114–2120
Mazanka E, Alexander J, Yeh BJ, Charoenpong P, Lowery DM, Yaffe M, Weiss EL (2008) The NDR/LATS family kinase Cbk1 directly controls transcriptional asymmetry. PLoS Biol 6:203
McCarty TP, Pappas PG (2016) Invasive candidiasis. Infect Dis Clin N Am 30:103–124
Min K, Ichikawa Y, Woolford CA, Mitchell AP (2016) Candida albicans gene deletion with a transient CRISPR-Cas9 system. mSphere 1:e00130-e216
Mulhern SM, Logue ME, Butler G (2006) Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot Cell 5:2001–2013
Nelson B, Kurischko C, Horecka J, Mody M, Nair P, Pratt L, Zougman A, McBroom LD, Hughes TR, Boone C, Luca FC (2003) RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol Biol Cell 14:3782–3803
Nett JE, Lepak AJ, Marchillo K, Andes DR (2009) Time course global gene expression analysis of an in vivo Candida biofilm. J Infect Dis 200:307–313
Noble SM, Johnson AD (2005) Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4:298–309
Romo JA, Kumamoto CA (2020) On commensalism of Candida. J Fungi 6:16
Saputo S, Chabrier-Rosello Y, Luca FC, Kumar A, Krysan DJ (2012) The RAM pathway in pathogenic fungi. Eukaryot Cell 11:708–717
Saputo S, Kumar A, Krysan DJ (2014) Efg1 directly regulates ACE2 expression to mediate cross talk between cAMP/PKA and RAM pathways during Candida albicans morphogenesis. Eukaryot Cell 13:1169–1180
Sbia M, Parnell EJ, Yu Y, Olsen AE, Kretschmann KL, Voth WP, Stillman DJ (2008) Regulation of the yeast Ace2 transcription factor during the cell cycle. J Biol Chem 283:11135–11145
Shen J, Cowen LE, Griffin AM, Chan L, Köhler JR (2008) The Candida albicans pescadillo homolog is required for normal hypha-to-yeast morphogenesis and yeast proliferation. Proc Natl Acad Sci USA 105:20918–20923
Sudbery PE (2011) Growth of Candida albicans hyphae. Nat Rev Microbiol 9:737–748
Uppuluri P, Chaturvedi AK, Srinivasan A, Banerjee M, Ramasubramaniam AK, Köhler JR, Kadosh D, Lopez-Ribot JL (2010) Dispersion as an important step in the Candida albicans biofilm development cycle. PLoS Pathog 26:e1000828
Wakade RS, Ristow LC, Stamnes MA, Kumar A, Krysan DJ (2020) The Ndr/LATS kinase Cbk1 regulates a specific subset of Ace2 function and suppresses the hypha-to-yeast transition in Candida albicans. mBio 11:e01900-e1920
Wall G, Montelongo-Jauregui D, Vidal Bonifacio B, Lopez-Ribot JL, Uppuluri P (2019) Candida albicans biofilm growth and dispersal: contribution to pathogenesis. Curr Opin Microbol 2:1–6
Acknowledgements
This work was supported by NIH grant 1R01AI133409 (DJK). We thank Aaron Mitchell (Georgia), Scott Filler (UCLA) and Scott Moye-Rowley (Iowa) for stimulating discussions regarding this project.
Author information
Authors and Affiliations
Contributions
RSW: performed experiments, composed figures, edited manuscript; DJK: prepared and edited manuscript, edited figures, and obtained funding.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests. All new data are reported in the text of the manuscript. Strains are available by contacting the corresponding author.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wakade, R.S., Krysan, D.J. The Cbk1-Ace2 axis guides Candida albicans from yeast to hyphae and back again. Curr Genet 67, 461–469 (2021). https://doi.org/10.1007/s00294-020-01152-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-020-01152-1