Abstract
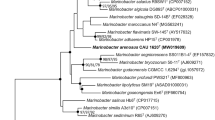
A bacterium designated GXH0434T was isolated from sea shore samples collected from Weizhou Island, Beihai, Guangxi, China. The organism is motile, strictly aerobic, and possesses a rod–coccus cell cycle in association with the growth phase. It can grow at 15–45 °C (optimum 37 °C), at pH 6.0–11.0 (optimum 6.0), and at 0–20% (w/v) NaCl (optimum 5.0–8.0%). The strain is positive for peroxidase and oxidase activity, negative for Voges-Proskauer test, can hydrolyze Tween 20, Tween 60, Tween 80, casein, and is able to produce siderophore and has the function of nitrogen fixation. Molecular phylogenetic analysis based on 16S rRNA gene sequences indicated that GXH0434T was most closely related to Microbulbifer halophilus KCTC 12848T with the similarity of 97.2%, followed by Microbulbifer chitinilyticus JCM 16148T (97.1%) and Microbulbifer taiwanensis LMG 26125T (96.5%). The digital DNA-DNA hybridization and the average nucleotide identity values between GXH0434T and Microbulbifer halophilus KCTC 12848T were 28.90% and 83.38%, respectively, which were below thresholds of species delineation. The genomic DNA G+C content of the strain was 61.9%. The major fatty acids were iso-C15:0, C16:0, iso-C11:0 3-OH, iso-C11:0 and Summed features 8 (C18:0 ω7c and/or C18:0 ω6c). The major polar lipids detected in GXH0434T were diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylcholine (PC). The major respiratory quinone was ubiquinone Q-8. Based on the above polyphasic classification indicated strain GXH0434T represents a novel species of the genus Microbulbifer, for which the name Microbulbifer litoralis sp. nov. is proposed. The type strain is GXH0434T (= MCCC 1K07158T = KCTC 92169T).

Similar content being viewed by others
References
González JM, Mayer F, Moran MA, Hodson RE, Whitman WB (1997) Microbulbifer hydrolyticus gen. nov., sp. nov., and Marinobacterium georgiense gen. nov., sp. nov., two marine bacteria from a lignin-rich pulp mill waste enrichment community. Int J Syst Bacteriol 47:369–376
Nishijima M, Takadera T, Imamura N, Kasai H, An KD, Adachi K, Nagao T, Sano H, Yamasato K (2009) Microbulbifer variabilis sp. nov. and Microbulbifer epialgicus sp. nov., isolated from Pacific marine algae, possess a rod-coccus cell cycle in association with the growth phase. Int J Syst Evol Microbiol 59(Pt 7):1696–1707. https://doi.org/10.1099/ijs.0.006452-0
Cheng Y, Zhu S, Guo C, Xie F, Jung D, Li S, Zhang W, He S (2021) Microbulbifer hainanensis sp. nov., a moderately halopilic bacterium isolated from mangrove sediment. Antonie Van Leeuwenhoek 114(7):1033–1042. https://doi.org/10.1007/s10482-021-01574-y
Vashist P, Nogi Y, Ghadi SC, Verma P, Shouche YS (2013) Microbulbifer mangrovi sp. Nov., a polysaccharide-degrading bacterium isolated from an Indian mangrove. Int J Syst Evol Microbiol 63(Pt 7):2532–2537. https://doi.org/10.1099/ijs.0.042978-0
Park S, Yoon SY, Ha MJ, Yoon JH (2017) Microbulbifer aestuariivivens sp. nov., isolated from a tidal flat. Int J Syst Evol Microbiol 67(5):1436–1441. https://doi.org/10.1099/ijsem.0.001831
Miyazaki M, Nogi Y, Ohta Y, Hatada Y, Fujiwara Y, Ito S, Horikoshi K (2008) Microbulbifer agarilyticus sp. nov. and Microbulbifer thermotolerans sp. Nov., agar-degrading bacteria isolated from deep-sea sediment. Int J Syst Evol Microbiol 58(Pt 5):1128–1133. https://doi.org/10.1099/ijs.0.65507-0
Zhang DS, Huo YY, Xu XW, Wu YH, Wang CS, Xu XF, Wu M (2012) Microbulbifer marinus sp. nov. and Microbulbifer yueqingensis sp. nov., isolated from marine sediment. Int J Syst Evol Microbiol 62(Pt 3):505–510. https://doi.org/10.1099/ijs.0.027714-0
Yoon JH, Kim IG, Oh TK, Park YH (2004) Microbulbifer maritimus sp. nov., isolated from an intertidal sediment from the Yellow Sea, Korea. Int J Syst Evol Microbiol 54(Pt 4):1111–1116. https://doi.org/10.1099/ijs.0.02985-0
Yoon JH, Kim IG, Shin DY, Kang KH, Park YH (2003) Microbulbifer salipaludis sp. Nov., a moderate halophile isolated from a Korean salt marsh. Int J Syst Evol Microbiol 53(Pt1):53–57. https://doi.org/10.1099/ijs.0.02342-0
Zhong W, Deutsch JM, Yi D, Abrahamse NH, Mohanty I, Moore SG, McShan AC, Garg N (2023) Discovery and biosynthesis of ureidopeptide natural products macrocyclized via indole n-acylation in marine Microbulbifer spp. Bacteria Chembiochem 24(12):e202300190. https://doi.org/10.1002/cbic.202300190
Li H, Huang X, Yao S, Zhang C, Hong X, Wu T, Jiang Z, Ni H, Zhu Y (2022) Characterization of a bifunctional and endolytic alginate lyase from Microbulbifer sp. ALW1 and its application in alginate oligosaccharides production from Laminaria japonica. Protein Expr Purif 200:106171. https://doi.org/10.1016/j.pep.2022.106171
Cho JY, Kim SH, Jung HJ, Cho DH, Kim BC, Bhatia SK, Ahn J, Jeon JM, Yoon JJ, Lee J, Yang YH (2022) Finding a benign plasticizer to enhance the microbial degradation of polyhydroxybutyrate (PHB) evaluated by PHB degrader Microbulbifer sp. SOL66. Polymers (Basel). 14(17):3625. https://doi.org/10.3390/polym14173625
de Lamballerie X, Zandotti C, Vignoli C, Bollet C, de Micco P (1992) A one-step microbial DNA extraction method using Chelex 100 suitable for gene amplification. Res Microbiol 143:785–790. https://doi.org/10.1016/0923-2508(92)90107-Y
Chun J, Goodfellow M (1995) A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int J Syst Bacteriol 45(2):240–245. https://doi.org/10.1099/00207713-45-2-240
Yoon SH, Kwon S, Lim J, Kim Y (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole- genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Tamura K, Stecher G, Kumar S (2021) MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol Biol Evol 38(7):3022–3027. https://doi.org/10.1093/molbev/msab120
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol 20:406–416. https://doi.org/10.1093/sysbio/20.4.406
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120. https://doi.org/10.1007/BF01731581
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.2307/2408678
Chen S, Zhou Y, Chen Y, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34(17):i884–i890. https://doi.org/10.1093/bioinformatics/bty560
Wick RR, Judd LM, Gorrie CL (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13(6):e1005595. https://doi.org/10.1371/journal.pcbi.1005595
Meier-Kolthoff JP, Sardà Carbasse J, Peinado-Olarte RL, Göker M (2022) TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acid Res 50:D801–D807. https://doi.org/10.1093/nar/gkab902
Meier-Kolthoff JP, Auch AF (2013) Klenk, HP (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. https://doi.org/10.1186/1471-2105-14-60
Yoon SH, Ha SM, Lim JM, Kwon SJ, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP et al (2021) AntiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49(1):29–35
Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T et al (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45:D535–D542
Brown AE (2007) Benson’s microbiological application laboratory manual in general microbiology, 10th edn. McGraw-Hill, New York
Gonzalez C, Gutierrez C, Ramirez C (1978) Halobacterium vallismortis sp. nov. an amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 24:710–715. https://doi.org/10.1139/m78-119
Collins MD (1977) Distrbution of menaquinone in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230. https://doi.org/10.1099/00221287-100-2-221
MinnikinO’donnell DA, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett J (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241. https://doi.org/10.1016/0167-7012(84)90018-6
Sasser M, Kunitsky C, Jackoway G, Ezzell JW, Teska JD (2005) Identification of Bacillus anthracis from culture using gas chromatographic analysis of fatty acid methyl esters. J AOAC Int 88:178–181. https://doi.org/10.1093/jaoac/88.1.178
Tang SK, Wang Y, Cai M, Lou K, Mao PH, Jin X, Jiang CL, Xu LH, Li WJ (2008) Microbulbifer halophilus sp. nov., a moderately halophilic bacterium from north-west China. Int J Syst Evol Microbiol 58(9):2036–2040. https://doi.org/10.1099/ijs.0.65519-0
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461. https://doi.org/10.1099/ijsem.0.002516
Liu R, Zhang Y, Chen P, Lin H, Ye G, Wang Z, Ge C, Zhu B, Ren D (2017) Genomic and phenotypic analyses of Pseudomonas psychrotolerans PRS08–11306 reveal a turnerbactin biosynthesis gene cluster that contributes to nitrogen fixation. J Biotechnol 253:10–13. https://doi.org/10.1016/j.jbiotec.2017.05.012
Acknowledgements
We thank the Korean Collection of Type Cultures (KCTC) for providing strain Microbulbifer halophilus KCTC 12848T, and Marine Culture Collection of China (MCCC) and Korean Collection for Type Cultures (KCTC) for strain deposited, as well as members of the Guangxi Key Laboratory of Polysaccharide Materials and Modifications and Yunnan Institute of Microbiology of Yunnan University for helpful discussions.
Funding
This research was funded by the Science and Technology Major Project of Guangxi (AA18242026) and Guangxi University Young and Mid-aged Teachers’ Basic Scientific Research Ability Improvement Project of Universities in Guangxi, China (2021KY0175).
Author information
Authors and Affiliations
Contributions
MJ and YJ designed the study; YH, YJ, AZ and XC performed research; YL, FW and HL analyzed data. YNI and WH resource; YH wrote the manuscript. All of the authors contributed to the manuscript revision process and read and approved the submitted version.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interest.
Ethical Approval
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene and the genome sequences of strain GXH0434T are OL336515 and JALKCV000000000, respectively. Strain GXH0434T has been deposited in Marine Culture Collection of China (MCCC) and Korean Collection for Type Cultures (KCTC), the preservation number are MCCC 1K07158 and KCTC 92169, respectively.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, Y., Jiang, Y., Zhao, A. et al. Microbulbifer litoralis sp. nov., Isolated from Seashore of Weizhou Island. Curr Microbiol 81, 105 (2024). https://doi.org/10.1007/s00284-023-03594-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03594-5