Abstract
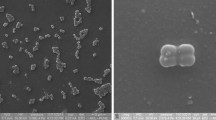
A Gram-staining-positive, non-motile, aerobic, spherical actinobacterium, designated WL0053T, was isolated from the coastal sediment of Nantong City, Jiangsu Province, China. The 16S rRNA gene sequence of strain WL0053T exhibited the highest similarities to Nocardioides mesophilus MSL-22T (98.0%), N. massiliensis GD12T (97.8%), Marmoricola bigeumensis MSL-05T (97.6%), and N. jensenii DSM 20641T (97.3%). The polyphasic taxonomic approach was used for the identification of strain WL0053T. This strain formed white, round, and smooth colonies and grew in the presence of 0–18% (w/v) NaCl (optimum, 0–4.0%), at pH 6.0–9.0 (optimum, pH 7.0) and at 20–37 °C (optimum, 28 °C). The main cellular fatty acids comprised of C17:1 ω8c, C18:1 ω9c, and iso-C16:0. The genomic DNA G + C content was 71.9%. The predominant quinone was MK-8(H4), and the major polar lipid consisted of phosphatidylcholine, glycolipid, phosphatidylethanolamine, and two unidentified phospholipids. Phylogenetic trees of 16S rRNA gene and bac120 gene set indicted that strain WL0053T was closely related to the species N. iriomotensis and N. mesophilus, while these two species clustered in a separate clade together with M. caldifontis YIM 730233T in the bac120 tree. Combined with the analysis of average nucleotide identity (ANI), average amino acid identity (AAI), and digital DNA–DNA hybridization (dDDH), it can be considered that the strain WL0053T is a new member of the genus Nocardioides and is proposed to be named as Nocardioides jiangsuensis sp. nov.. The type strain is WL0053T (=MCCC 1K05897T=JCM 34671T=GDMCC 4.192T). Furthermore, based on the fact that the genera Nocardioides and Marmoricola both appeared polyphyletic with no significant difference on phenotypic and chemotaxonomic traits, we proposed to reclassify the genus Marmoricola as Nocardioides.


Similar content being viewed by others
Data Availability
All of the data supporting the conclusions of this article are included within the article and its additional files. The genome datasets and the 16S rRNA gene sequence of Nocardioides jiangsuensis WL0053T generated during the current study are available in the GenBank repository under accession number JAIEZQ000000000 and OK473545, respectively. Other datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Code Availability
Not applicable.
References
Parte AC (2018) LPSN—list of prokaryotic names with standing in nomenclature (bacterio.net), 20 years on. Int J Syst Evol Microbiol 68:1825–1829. https://doi.org/10.1099/ijsem.0.002786
Lee SD (2007) Marmoricola aequoreus sp. nov., a novel actinobacterium isolated from marine sediment. Int J Syst Evol Microbiol 57:1391–1395. https://doi.org/10.1099/ijs.0.64696-0
Kim SJ et al (2015) Marmoricola solisilvae sp. nov. and Marmoricola terrae sp. nov., I solated from soil and emended description of the genus Marmoricola. Int J Syst Evol Microbiol 65:1825–1830. https://doi.org/10.1099/ijs.0.000184
Jiang ZK et al (2017) Marmoricola endophyticus sp. nov., an endophytic actinobacterium isolated from Thespesia populnea. Int J Syst Evol Microbiol 67:4379–4384. https://doi.org/10.1099/ijsem.0.002297
Urzì C, Salamone P, Schumann P, Stackebrandt E (2000) Marmoricola aurantiacus gen. nov., sp. nov., a coccoid member of the family Nocardioidaceae isolated from a marble statue. Int J Syst Evol Microbiol 50:529–536. https://doi.org/10.1099/00207713-50-2-529
Lee DW, Lee SD (2010) Marmoricola scoriae sp. nov., isolated from volcanic ash. Int J Syst Evol Microbiol 60:2135–2139. https://doi.org/10.1099/ijs.0.018242-0
Habib N et al (2020) Marmoricola caldifontis sp. nov., a novel actinobacterium isolated from a hot spring. Int J Syst Evol Microbiol 70:2053–2058. https://doi.org/10.1099/ijsem.0.004016
De Menezes CBA et al (2015) Marmoricola aquaticus sp. nov., an actinomycete isolated from a marine sponge. Int J Syst Evol Microbiol 65:2286–2291. https://doi.org/10.1099/ijs.0.000254
Jang JH, Maeng S, Jung H-Y, Kim MK, Subramani G (2020) Nocardioides jejuensis sp. nov., isolated from soil. Antonie Van Leeuwenhoek 113:339–347. https://doi.org/10.1007/s10482-019-01343-y
Prauser H (1976) Nocardioides, a new genus of the order Actinomycetales. Int Assoc Microbiol Soc 26(1):58–65. https://doi.org/10.1099/00207713-26-1-58
O’Donnell AG, Goodfellow M, Minnikin DE (1982) Lipids in the classification of Nocardioides: reclassification of Arthrobacter simplex (Jensen) lochhead in the genus Nocardioides (Prauser) emend. O’Donnell et al. as Nocardioides simplex comb. nov. Arch Microbiol 133(4):323–329. https://doi.org/10.1007/BF00521299
Park Y et al (2020) Nocardioides convexus sp. nov. and Nocardioides anomalus sp. nov., isolated from soil and mineral water. Int J Syst Evol Microbiol 70(12):6402–6407. https://doi.org/10.1099/ijsem.0.004547
Schumann P et al (2018) Marmoricola silvestris sp. nov., a novel actinobacterium isolated from alpine forest soil. Int J Syst Evol Microbiol 68(4):1313–1318. https://doi.org/10.1099/ijsem.0.002674
Zhang D-F et al (2014) Nocardioides nanhaiensis sp. nov., an actinobacterium isolated from a marine sediment sample. Int J Syst Evol Microbiol 64:2718–2722. https://doi.org/10.1099/ijs.0.062851-0
Lee L-H, Zainal N, Azman A-S, Mutalib N-SA, Hong K, Chan K-G (2014) Mumia flava gen. nov., sp. nov., an actinobacterium of the family Nocardioidaceae. Int J Syst Evol Microbiol 64:1461–1467. https://doi.org/10.1099/ijs.0.058701-0
Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, Hugenholtz P (2018) A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. https://doi.org/10.1038/nbt.4229
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. https://doi.org/10.1128/jb.173.2.697-703.1991
Richter M, Rossello-Mora R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131. https://doi.org/10.1073/pnas.0906412106
Nurk S, Bankevich A, Antipov D et al (2013) Assembling single-cell genomes and minimetagenomes from chimeric MDA products. J Comput Biol 20:714–737. https://doi.org/10.1089/cmb.2013.0084
Okonechnikov K, Golosova O, Fursov M, team tU (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. https://doi.org/10.1093/bioinformatics/bts091
Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60. https://doi.org/10.1186/1471-2105-14-60
Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, Zhou J, Oren A, Zhang YZ (2014) A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196:2210–2215. https://doi.org/10.1128/jb.01688-14
Tatusova T et al (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. https://doi.org/10.1093/nar/gkw569
Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P (2017) Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol Biol Evol 34:2115–2122. https://doi.org/10.1093/molbev/msx148
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. https://doi.org/10.1093/nar/gkv1070
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol 20:406–416. https://doi.org/10.2307/2412116
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Zhang D-F, Cui X-W, Zhao Z, Zhang A-H, Huang J-K, Li W-J (2020) Sphingomonas hominis sp. nov., isolated from hair of a 21-year-old girl. Antonie Van Leeuwenhoek 113:1523–1530. https://doi.org/10.1007/s10482-020-01460-z
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI technical note 101. Microbial ID, Inc, Newark
Tamaoka J (1986) Analysis of bacterial menaquinone mixtures by reverse-phase high-performance liquid chromatography. Methods Enzymol 123:251–256. https://doi.org/10.1016/s0076-6879(86)23028-1
Collins MD, Jones D (1981) A note on the separation of natural mixtures of bacterial ubiquinones using reverse-phase partition thin-layer chromatography and high performance liquid chromatography. J Appl Bacteriol 51:129–134. https://doi.org/10.1111/j.1365-2672.1981.tb00916.x
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241. https://doi.org/10.1016/0167-7012(84)90018-6
Mitchell AL et al (2020) MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res 48:D570–D578. https://doi.org/10.1093/nar/gkz1035
Sattin SR et al (2009) Functional shifts in unvegetated, perhumid, recently-deglaciated soils do not correlate with shifts in soil bacterial community composition. J Microbiol 47:673–681. https://doi.org/10.1007/s12275-009-0194-7
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. https://doi.org/10.1099/ijs.0.64483-0
Dastager SG et al (2008) Marmoricola bigeumensis sp. nov., a member of the family Nocardioidaceae. Int J Syst Evol Microbiol 58(5):1060–1063. https://doi.org/10.1099/ijs.0.65576-0
Lee H-Y et al (2016) Marmoricola ginsengisoli sp. nov. and Marmoricola pocheonensis sp. nov. isolated from a ginseng-cultivating field. Int J Syst Evol Microbiol 66(5):1996–2001. https://doi.org/10.1099/ijsem.0.000977
Parker CT, Tindall BJ, Garrity GM (2019) International code of nomenclature of prokaryotes. Int J Syst Evol Microbiol 69(1A):S1–S111. https://doi.org/10.1099/ijsem.0.000778
Lin SY et al (2015) Nocardioides echinoideorum sp. nov., isolated from sea urchins (Tripneustes gratilla). Int J Syst Evol Microbiol 65(6):1953–1958. https://doi.org/10.1099/ijs.0.000206
Acknowledgements
We are grateful to Professor Aharon Oren (The Hebrew University of Jerusalem, Israel) and Professor Wen-Jun Li (Sun Yat-Sen University, China) for the Latin construction of the taxa names.
Funding
This research was supported by the National Natural Science Foundation of China (No. 31900001), the China Postdoctoral Science Foundation (2020M671312) and the Fundamental Research Funds for the Central Universities (B210202140).
Author information
Authors and Affiliations
Contributions
D-FZ and LW designed research and project outline. LW and D-FZ performed isolation, deposition, and polyphasic taxonomy. D-FZ, LW, and H-PX performed genome analysis. LW, D-FZ, and AZ drafted the manuscript. J-KH and CL revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to Participate
All the authors listed have approved this manuscript and agreed to authorship and submission of the manuscript for peer review.
Consent for Publication
All the authors listed have approved this manuscript and agreed to publication on Current Microbiology.
Declaration of Deposition in Repositories
The type strain Nocardioides jiangsuensis WL0053T had been deposited in Marine Culture Collection of China (MCCC), Japan Collection of Microorganisms (JCM), and Guangdong Microbial Culture Collection Center (GDMCC).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, L., Xue, HP., Zhang, DF. et al. Description of Nocardioides jiangsuensis sp. nov., and Proposal for Reclassification of the Genus Marmoricola as Nocardioides. Curr Microbiol 80, 60 (2023). https://doi.org/10.1007/s00284-022-02977-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-02977-4