Abstract
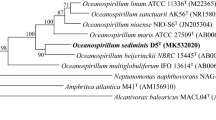
A Gram-stain-negative, rod-shaped, motile, mesophilic, and aerobic bacterial strain, designated SM2-42 T was isolated from a mangrove sediment. Catalase activity and oxidase activity were positive. Growth was observed at 20 °C–40 °C, pH 6.0–8.0, and in the presence of 0.5–5.0% NaCl. Cells of strain SM2-42 T contained poly-β-hydroxybutyrate granules. The 16S rRNA gene of strain SM2-42 T had maximum sequence similarity with Oceanobacter kriegii 197 T of 97.1%. Phylogenetic analysis based on 16S rRNA gene sequence and 120 conserved concatenated proteins indicated that strain SM2-42 T was affiliated to the genus Oceanobacter and formed a monophyletic branch with O. kriegii 197 T. The average nucleotide identity and digital DNA-DNA hybridization values between strain SM2-42 T and O. kriegii 197 T were 76.43% and 21.60%, respectively. The major isoprenoid quinone was Q-8. The major fatty acids (> 10%) comprised C16:0, summed feature 8 (C18:1ω7c and C18:1 ω6c), C18:0, and summed feature 3 (C16:1ω7c and/or C16:1 ω6c). The polar lipid profile consisted of phosphatidylethanolamine, phosphatidylglycerol, one unidentified aminolipid and two unidentified lipids. The draft genome size was 5,115,008 bp with DNA G + C content of 54.3%. Based on phylogenetic analyses and whole genomic comparisons, strain SM2-42 T represented a novel species, for which the name Oceanobacter mangrovi sp. nov. was proposed. The type strain was SM2-42 T (= MCCC 1K06300T = KCTC 82938 T).


Similar content being viewed by others
Data Availability
The GenBank/EMBL/DDBJ accession numbers of the 16S rRNA gene sequence and the draft genome sequence of strain SM2-42 T are MT829535 and JAHXZR000000000, respectively.
Abbreviations
- MCCC:
-
Marine culture collection of China
- KCTC:
-
Korean collection for type cultures
- ANI:
-
Average nucleotide identity
- dDDH:
-
Digital DNA-DNA hybridization
References
Satomi M, Kimura B, Hamada T, Harayama S, Fujii T (2002) Phylogenetic study of the genus Oceanospirillum based on 16S rRNA and gyrB genes: emended description of the genus Oceanospirillum, description of Pseudospirillum gen. nov., Oceanobacter gen. nov. and Terasakiella gen. nov. and transfer of Oceanospirillum jannaschii and Pseudomonas stanieri to Marinobacterium as Marinobacterium jannaschii comb. nov. and Marinobacterium stanieri comb. no. Int J Syst Evol Microbiol 52(3):739–747. https://doi.org/10.1099/00207713-52-3-739
Baumann L, Baumann P, Mandel M, Allen RD (1972) Taxonomy of aerobic marine eubacteria. J Bacteriol 110(1):402–429. https://doi.org/10.1128/jb.110.1.402-429.1972
Teramoto M, Ohuchi M, Hatmanti A, Darmayati Y, Widyastuti Y, Harayama S, Fukunaga Y (2011) Oleibacter marinus gen. nov., sp. nov., a bacterium that degrades petroleum aliphatic hydrocarbons in a tropical marine environment. Int J Syst Evol Microbiol 61(2):375–380. https://doi.org/10.1099/ijs.0.018671-0
Teramoto M, Suzuki M, Okazaki F, Hatmanti A, Harayama S (2009) Oceanobacter-related bacteria are important for the degradation of petroleum aliphatic hydrocarbons in the tropical marine environment. Microbiology (Reading) 155(10):3362–3370. https://doi.org/10.1099/mic.0.030411-0
Huang Z, Mo S, Yan L, Wei X, Huang Y, Zhang L, Zhang S, Liu J, Xiao Q, Lin H, Guo Y (2021) A simple culture method enhances the recovery of culturable actinobacteria from coastal sediments. Front Microbiol 12:675048–675051. https://doi.org/10.3389/fmicb.2021.675048
Huang Z, Hu Y, Lai Q, Guo Y (2020) Description of Maribellus sediminis sp. nov., a marine nitrogen-fixing bacterium isolated from sediment of cordgrass and mangrove. Syst Appl Microbiol 43(4):126099. https://doi.org/10.1016/j.syapm.2020.126099
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67(5):1613–1617. https://doi.org/10.1099/ijsem.0.001755
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477. https://doi.org/10.1089/cmb.2012.0021
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. https://doi.org/10.1186/1471-2105-11-119
Cantalapiedra CP, Hernandez-Plaza A, Letunic I, Bork P, Huerta-Cepas J (2021) eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol. https://doi.org/10.1093/molbev/msab293
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:1–14. https://doi.org/10.1186/1471-2105-14-60
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110(10):1281–1286. https://doi.org/10.1007/s10482-017-0844-4
Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P (2018) A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36(10):996–1004. https://doi.org/10.1038/nbt.4229
Letunic I, Bork P (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23(1):127–128. https://doi.org/10.1093/bioinformatics/btl529
Huang Z, Dong C, Shao Z (2016) Paraphotobacterium marinum gen. nov., sp. nov., a new member of the family Vibrionaceae, isolated from the surface seawater of the South China Sea. Int J Syst Evol Microbiol 66:3050–3056. https://doi.org/10.1099/ijsem.0.001142
Komagata K, Suzuki K (1987) Lipid and cell-wall analysis in bacterial systematics. Methods in Microbiology 19:161–207. https://doi.org/10.1016/S0580-9517(08)70410-0
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, De Meyer S, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68(1):461–466. https://doi.org/10.1099/ijsem.0.002516
Bowditch RD, Baumann L, Baumann P (1984) Description of Oceanospirillum kriegii sp. nov. and O. jannaschii sp. nov. and assignment of two species of Alteromonas to this genus as O. commune comb. nov. and O. vagum comb. nov. Curr Microbiol 10:221–230
Funding
This work was supported by the project of Natural Science Foundation of Fujian Province (2020J01789) and National Science Foundation of China (41706139).
Author information
Authors and Affiliations
Contributions
ZH conceived the study and drafted the manuscript. ZH, YH, QL, XC, CD and XH performed the experiment and analyze the data.
Corresponding author
Ethics declarations
Conflict of Interest
No conflicts of interest are declared by the authors.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the author.
Consent for Publication
The authors approved for the publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, Z., Huang, Y., Lai, Q. et al. Oceanobacter mangrovi Sp. Nov., a Novel Poly-β-hydroxybutyrate Accumulating Bacterium Isolated from Mangrove Sediment. Curr Microbiol 79, 100 (2022). https://doi.org/10.1007/s00284-022-02798-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-02798-5