Abstract
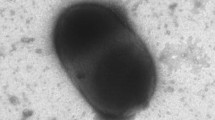
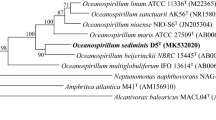
A Gram-negative bacterium, denoted JLT2012T, was isolated from the surface water of the Pacific Ocean. This aerobic bacterium was rod shaped and devoid of flagella, displayed gliding motility, and grew in characteristic orange colonies. The bacterium contained ubiquinone Q-10 as the major respiratory quinone, and spermidine and spermine as the major polyamine compounds. The dominant fatty acids were C18:1ω7c and/or C18:1ω6c (34.7 %), C16:0 (21.3 %), and C18:0 (15.9 %), whereas the polar lipids consisted mainly of diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, four sphingoglycolipids, and several unknown glycolipids. The G + C content DNA was found to be 65.5 mol%. Comparative 16S rRNA gene sequence analysis revealed that strain JLT2012T formed a distinct lineage within the genus Pacificimonas (formerly known as Pacificamonas) and shared the highest sequence similarity with the type strain of Pacificimonas flava JLT2015T (96.0 %). Data combined from different studies on the phenotypic, phylogenetic, and genomic characteristics indicated that strain JLT2012T is a representative of a novel species within Pacificimonas for which the name Pacificimonas aurantium sp. nov. (type strain JLT2012T=LMG 27361T=CGMCC 1.12399T) is proposed.

Similar content being viewed by others
References
Busse J, Auling G (1988) Polyamine pattern as a chemotaxonomic marker within the Proteobacteria. Syst Appl Microbiol 11:1–8
Collins MD, Goodfellow M, Minnikin DE (1980) Fatty acid, isoprenoid quinone and polar lipid composition in the classification of Curtobacterium and related taxa. J Gen Microbiol 118:29–37
Dong XZ, Cai MY (2001) Determinative manual for routine bacteriology (English translation). Scientific Press, Beijing, pp 370–390
Embley TM (1991) The linear PCR reaction: a simple and robust method for sequencing amplified rRNA genes. Lett Appl Microbiol 13:171–174
Gerhardt P, Murray RGE, Wood WA, Krieg NR (1994) Methods for general and molecular bacteriology. American Society for Microbiology, Washington
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Kates M (1986) Techniques of lipidology: isolation, analysis and identification of lipids: laboratory techniques in biochemistry and molecular biology. Biochemistry 25:285
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kim SB, Falconer C, Williams E, Goodfellow M (1998) Streptomyces thermocarboxydovorans sp. nov. and Streptomyces thermocarboxydus sp. nov., two moderately thermophilic carboxydotrophic species from soil. Int J Syst Bacteriol 48:59–68
Komagata K, Suzuki KI (1988) 4 Lipid and cell-wall analysis in bacterial systematics. Method Microbiol 19:161–207
Kosako Y, Yabuuchi E, Naka T, Fujiwara N, Kobayashi K (2000) Proposal of Sphingomonadaceae fam. nov., consisting of Sphingomonas Yabuuchi et al. 1990, Erythrobacter Shiba and Shimidu 1982, Erythromicrobium Yurkov et al. 1994, Porphyrobacter Fuerst et al. 1993, Zymomonas Kluyver and van Niel 1936, and Sandaracinobacter Yurkov et al. 1997, with the type genus Sphingomonas Yabuuchi et al. 1990. Microbiol Immunol 44:563–575
Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, DeMaere MZ, Ting L, Ertan H, Johnson J (2009) The genomic basis of trophic strategy in marine bacteria. PNAS 106:15527–15533
Liu K, Li S, Jiao N, Tang K (2014) Pacificamonas flava gen. nov., sp. nov., a novel member of the family Sphingomonadaceae isolated from the southeastern pacific. Curr Microbiol 69:96–101
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol 3:208-IN201
Maruyama T, Park HD, Ozawa K, Tanaka Y, Sumino T, Hamana K, Hiraishi A, Kato K (2006) Sphingosinicella microcystinivorans gen. nov., sp. nov., a microcystin-degrading bacterium. Int J Syst Evol Microbiol 56:85–89
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G + C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Ostle AG, Holt J (1982) Nile blue A as a fluorescent stain for poly-beta-hydroxybutyrate. Appl Environ Microbiol 44:238–241
Rüger HJ, Krambeck HJ (1994) Evaluation of the BIOLOG substrate metabolism system for classification of marine bacteria. Syst Appl Microbiol 17:281–288
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Smibert R, Krieg N (1994) phenotypic characterization: in methods for general and molecular bacteriology. American Society for Microbiology, Washington
Sowell SM, Wilhelm LJ, Norbeck AD et al (2009) Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J 3:93–105
Stingl U, Desiderio RA, Cho JC, Vergin KL, Giovannoni SJ (2007) The SAR92 clade: an abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl Environ Microbiol 73:2290–2296
Sun J, Steindler L, Thrash JC, Halsey KH, Smith DP, Carter AE, Landry ZC, Giovannoni SJ (2011) One carbon metabolism in SAR11 pelagic marine bacteria. PLoS One 6:e23973
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Takeuchi M, Hamana K, Hiraishi A (2001) Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int J Syst Evol Microbiol 51:1405–1417
Tang K, Jiao N, Liu K, Zhang Y, Li S (2012) Distribution and functions of TonB-dependent transporters in marine bacteria and environments: implications for dissolved organic matter utilization. PLoS One 7:e41204
Tindall B (1990) A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst Appl Microbiol 13:128–130
Tindall B (1990) Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol Lett 66:199–202
Acknowledgments
This work was supported by the National Natural Science Foundation of China Project (41276131), the Natural Science Foundation of Fujian Province (2014J01164), and the Fundamental Research Funds for the Central Universities (20720150078) (CNOOC-KJ 125 FZDXM 00 ZJ 001-2014).
Author information
Authors and Affiliations
Corresponding author
Additional information
The GenBank accession number for the 16S rRNA gene sequence of strain JLT2012T is JX878395.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, S., Zhou, W., Lin, D. et al. Pacificimonas aurantium sp. nov., Isolated from the Seawater of the Pacific Ocean. Curr Microbiol 72, 752–757 (2016). https://doi.org/10.1007/s00284-016-0999-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-016-0999-7